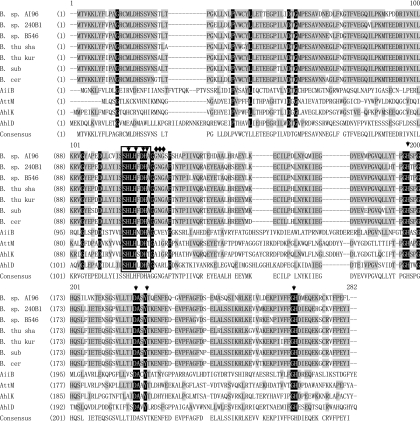
Fig 1.
Amino acid sequence alignment of AiiAAI96 with Bacillus AiiA-like proteins and with AHL lactonases from other species. The sequences are those of Bacillus sp. AI96 CGMCC 4164 (GenBank accession number HM750247) (B. sp. AI96), Bacillus sp. strain 240B1 (accession number AAF62398) (B. sp. 240B1), Bacillus sp. strain B546 (accession number FJ816104) (B. sp. B546), Bacillus thuringiensis serovar shandongiensis (accession number AAR85481) (B. thu sha) (referred to as AiiA SS10 in this paper), Bacillus thuringiensis serovar kurstaki (accession number AAM92140) (B. thu kur) (referred to as PDB accession number 2A7M in Fig. 2), Bacillus cereus (accession number ACR46836) (B. cer), Bacillus subtilis (accession number AAY51611) (B. sub), Arthrobacter sp. strain IBN110 (accession number AAP57766) (AhlD), Klebsiella pneumoniae (accession number AAO47340) (AhlK), Agrobacterium tumefaciens C58 (accession number AAK91031) (AiiB), and Agrobacterium tumefaciens C58 (accession number AAD43990) (AttM). The similar and identical residues are shaded in gray and black, respectively. The conserved motif HXHXDH is boxed. The active-site amino acids are indicated with arrows, and the amino acids involved in the modification of a helix to a random coil (Fig. 2) are indicated with diamonds.