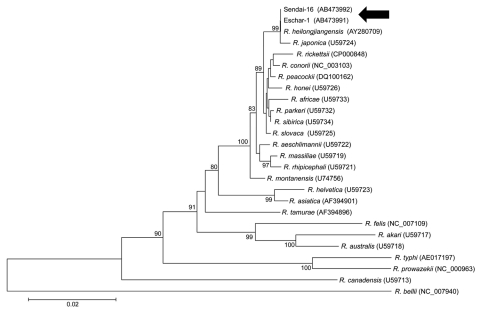
Figure 1.
Phylogenetic analysis of citrate synthase (gltA) sequences of Rickettsia spp. Sequences were aligned by using MEGA4 software (www.megasoftware.net). Neighbor-joining phylogenetic tree construction and bootstrap analyses were performed according to the Kimura 2-parameter distances method. Pairwise alignments and multiple alignments were performed with an open gap penalty of 15 and a gap extension penalty of 6.66. The percentage of replicate trees in which the associated taxa were clustered together in the bootstrap test (1,000 replicates) was calculated. Phylogenetic branches were supported by bootstrap values of >80%. All positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons (pairwise deletion). Scale bar indicates the percentage of sequence divergence. Arrows indicate eschar specimens.