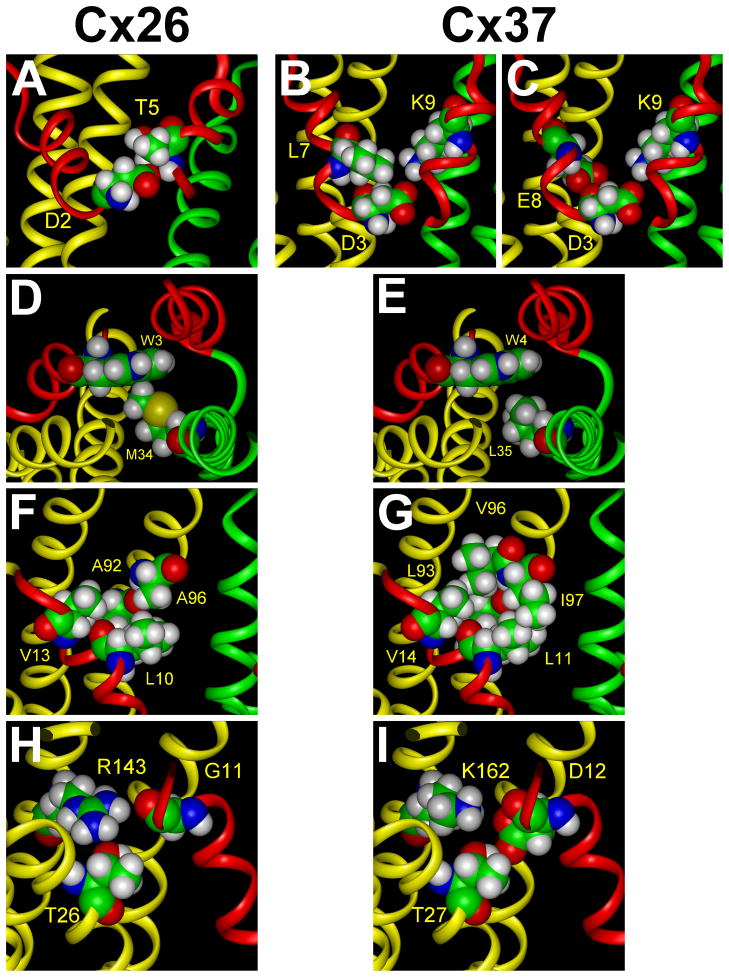
Figure 4.
Comparison of the interactions of Cx26 NT amino acids with those predicted for the Cx37 NT based on homology modeling. A. In Cx26, Asp2 forms a hydrogen bond with Thr5 of the adjacent monomer. B. In Cx37, Asp3 similarly forms a hydrogen bond with Phe6 of the adjacent monomer (not shown) and additionally participates in hydrophobic interactions with Leu7 in the same monomer. C. In Cx37, both Asp3 and Glu8 interact electrostatically with Lys9 of the adjacent subunit. D. In Cx26, the side chain of Trp3 participates in van der Waals contacts with Met34 in TM1 of the adjacent subunit. E. In Cx37, there is a weaker interaction between the corresponding amino acid residues, Trp4 and Leu35, because their side chains are farther apart. F. In Cx26, Leu10 participates in hydrophobic interactions with Ala92 and Ala96 in TM2 of the same subunit, and Val13 interacts with Ala92 and Val95 (not shown). G. In Cx37, Leu11 has more extensive interactions with the corresponding Leu93 and Ile97 because of their longer side chains. Val14 has significant hydrophobic interactions with Leu93 and Val96. H. In Cx26, Gly11 does not have significant electrostatic or hydrogen bonding interactions with Thr26 and Arg143 although their side chains are in proximity. I. In Cx37, Asp12 (the substitution for Gly11) interacts through a hydrogen bond with Thr27 in TM1 and electrostatically with Lys162 in TM3 of the same subunit.