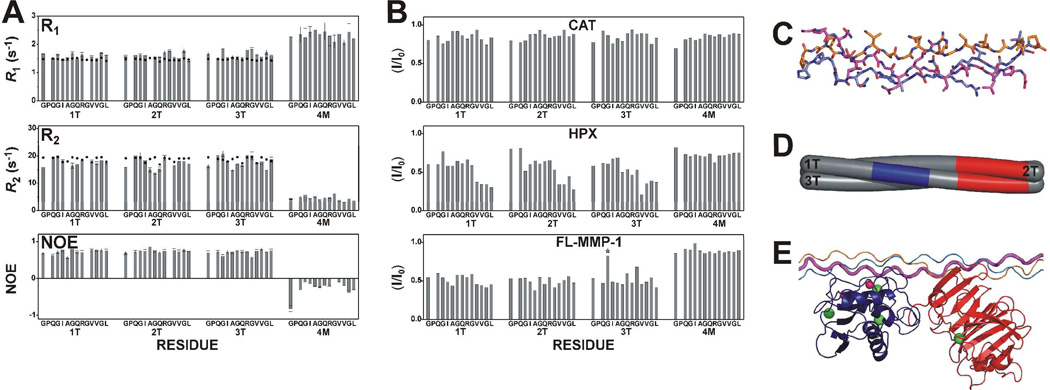
Figure 2.
(A) Experimental (bars) and theoretical (dots) R1, R2, and NOE values for rigid triple-helical (1T, 2T, and 3T) and non-triple-helical (4M) structure estimated from atomic coordinates of THPs using the program HYDRONMR and assuming for THP an S2 value of 0.80 at 298 K, typical for proteins. The THP structure was obtained by modeling the peptide on the crystal structure of an analogous THP (PDB code: 2d3h), and then refined with experimental NMR constraints. (B) Intensity changes per residue observed at 310 K in the 13C,15N THP upon the addition of unlabeled CAT (0.2 eq), HPX (0.2 eq), or FL-MMP-1 (0.2 eq). Residues showing signal overlap are indicated by stars. The only overlap is residue Gly16 from chain 3T with an impurity. (C) Solution structure of the THP. The three chains forming the THP are 1T (magenta), 2T (orange), and 3T (blue). (D) Schematic section of the THP with the stretch of chain 1T facing the active site of the CAT domain in blue and the HPX binding regions on chains 1T and 2T in red. (E) The adduct where the THP is still in a compact triple-helical conformation, with the CAT and HPX domains binding the cleavage site and the C-terminal region of the labeled stretch, respectively.