Figure 2.
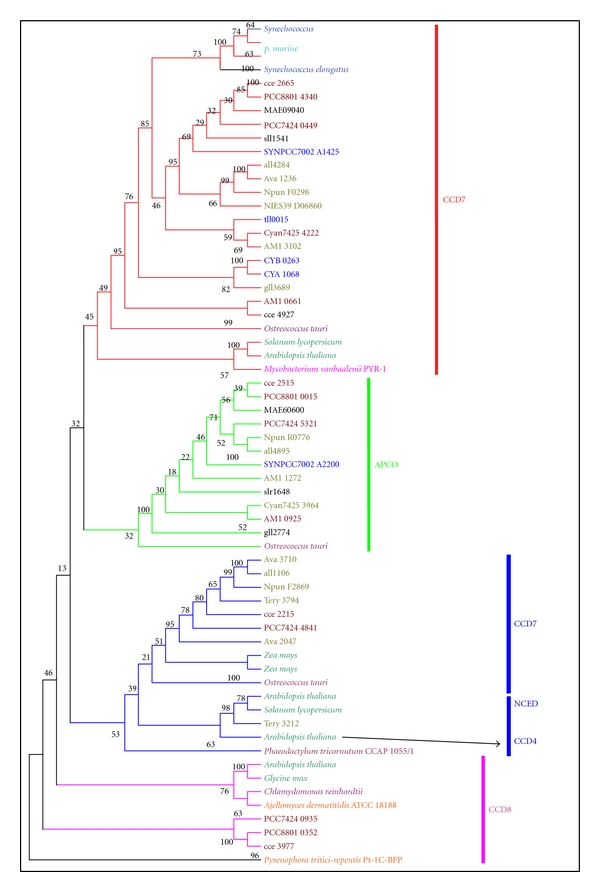
Maximum-likelihood tree of CCD enzymes of cyanobacteria, bacteria, eukaryotic algae, fungi, and higher plants. The model of LG+I+G was applied to construct ML tree using PHYML as described in the methods section. Numbers at the node indicated bootstrap values (%) for 1,000 replicates. Cyanobacterial CCD enzymes IDs and the strain names are as in Table 2 and Figure 1. Red line: CCD7, green: APCO, blue: CCD1, CCD4, and NCED, pink: CCD8. Species belonged to similar clade were represented by colored boxes. Orange: fungi, wheat: bacteria, purple: eukaryotic algae, barium: higher plants, cyan: cyanobacterial Prochlorococcus marinus, blue: Thermosynechococcus elongates, Synechococcus elongates, and Synechococcus, ruby: cyanobacterial Cyanothece, and smudge: filamentous cyanobacteria. Unlabeled species included Synechocystis sp. PCC 6803 (sll1541 and slr1648), Gloeobacter violaceus PCC 7421 (gll3689 and gll2774), Microcystis aeruginosa NIES-843 (MAE09040 and MAE60600).
