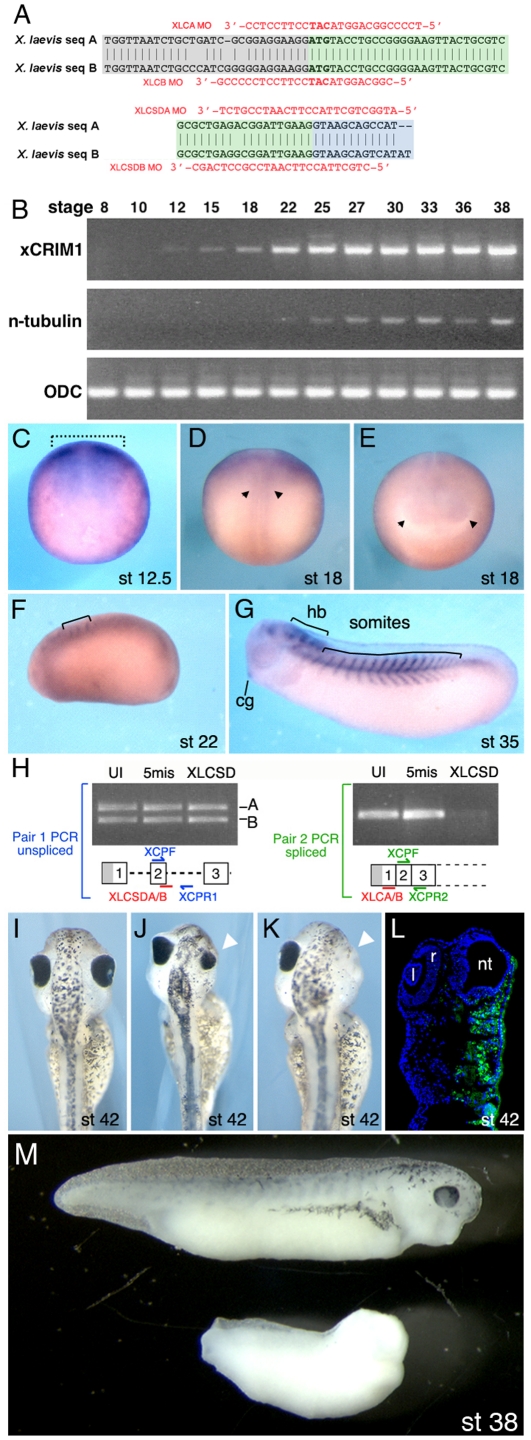
Figure 1. CRIM1 is expressed in the neural plate and is required for development of neural structures.
(A) Antisense morpholino oligo (MO) sequences and their complimentary targets on the Xenopus laevis messenger sequences are indicated for both the translation blocking MOs (upper) and the splice-donor blocking MOs (lower). (B) RT-PCR assessment of CRIM1A transcript expression in developmentally staged Xenopus laevis embryos. n-tubulin is a neuronal differentiation marker and ornithine decarboxylase (ODC) a ubiquitously expressed control. (C–G) In situ hybridizations for CRIM1A in Xenopus laevis embryos. The dashed bracket in (C) indicates the neural plate region, the arrowheads in (D) the neural tube and in (E) the optic vesicles. cg: cement gland; hb: hindbrain. (H) Detection of unspliced or spliced CRIM1A and B mRNA in uninjected embryos (UI) or those injected with the 5-missense control (5mis) or splice blocking (XLCSD) MOs. The PCR primer pair XCPF and XCPR1 detects unspliced mRNA while XCPF and XCPR2 detect the spliced product. (I–L) CRIM1 loss-of-function experiments where (I) is an uninjected control and (J–L) were injected with XLCA and B MOs at 15 ng each. Small (J, arrowhead) or missing eyes (K, arrowhead) often result. The embryo in (L) was coinjected with 10 kDa Alexa-fluor 488 dextran (green). l: lens; r: retina; nt: neural tube. (M) Comparison of control embryo (upper) and embryo injected at the 2-cell stage bi-laterally with XLCAB at 30 ng each (lower). Developmental stages as indicated.