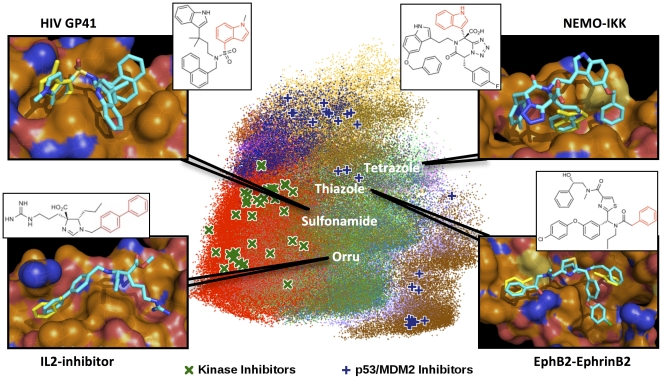
Figure 1. Representation of the chemical diversity of our multi-component reaction aromatic-biased libraries (different chemotypes shown in different colors) relative to the ZINC database [20] (red dots) and four predicted ligands.
The diversity space is visualized by plotting the top two principal components of the OpenBabel FP2 (http://openbabel.org) fingerprints of 200,000 compounds randomly selected from the 17.5-and-16 million compounds of ZINC and our aromatic-biased database, respectively. The PPI-biased compounds are focused on a different region of chemical space than the historically-biased ZINC database. Indeed, a library of kinase inhibitors, some containing a tryptophan analog, falls squarely in the space covered by ZINC, while inhibitors of p53/MDM2, including inhibitors without a tryptophan analog, are located in the space covered by the new libraries. Four novel compounds from four distinct scaffolds are found to match anchors on the GP41 dimer [38], IKK [39], IL-2 [40] and EphB2 [41] receptors. Complete reaction chemistries of the AnchorQuery libraries can be found at http://anchorquery.ccbb.pitt.edu.