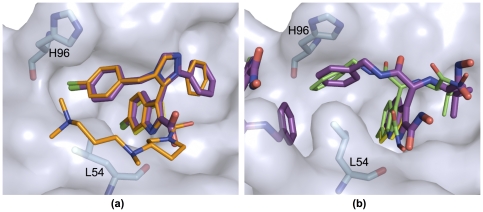
Figure 4. Two crystal structures of p53/MDM2 inhibitors validate the anchor-centric approach and docked models.
In both structures the indole anchor analog of tryptophan overlaps almost perfectly with W23 in p53 (shown in yellow sticks), when the receptors are aligned the MDM2 structure in the co-crystal (PDB 1YCR). (a) The ligand (purple sticks) of PDB 3LBK has a very similar binding mode to the number one hit in our virtual screen (orange sticks). (b) The crystal pose of the AnchorQuery derived compound (purple) with the predicted pose (green) also aligned very well. The presence of a second ligand near the binding interface distorts the receptor shape relative to the receptor used for docking.