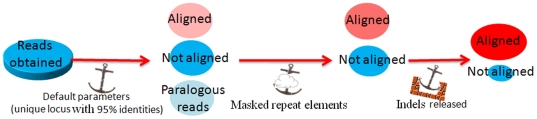
Figure 6. Summary of the alignment method used in the present study.
Alignment was accomplished in three successive steps: i) The first alignment used Mosaik with default parameters for 454 GS-FLX: 95% alignment homology in the sequences; ii) Reads not aligned in the first step and that were not paralogs were then filtered with RepeatMasker software. Reads with less than 90% homology with repeat elements were aligned by Mosaik with default parameters. iii) For reads not aligned in the second step, a third alignment was performed using a gap parameter fixed at a minimum (0.1 gap open and extensive penalties).