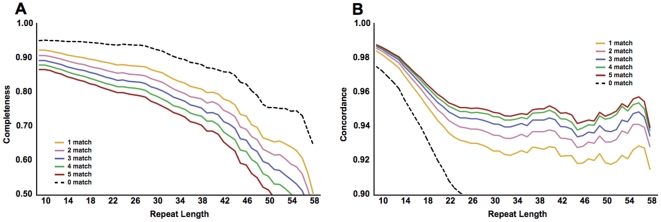
Figure 4. Increasing the requisite number of matching flank bases between a read and the reference improves genotyping accuracy at the expense of coverage.
The minimum required number of matching flanking bases for a read to be scored was incremented from zero to five. Two or more scorable reads were required to determine a repeat genotype. The plotted values are the mean (A) completeness (fraction of repeats with at least two reads passing filtering criteria) and (B) concordance for the genomes in the DGRP panel. Data have been smoothed for clarity (unweighted mean with window size ±2 bases).