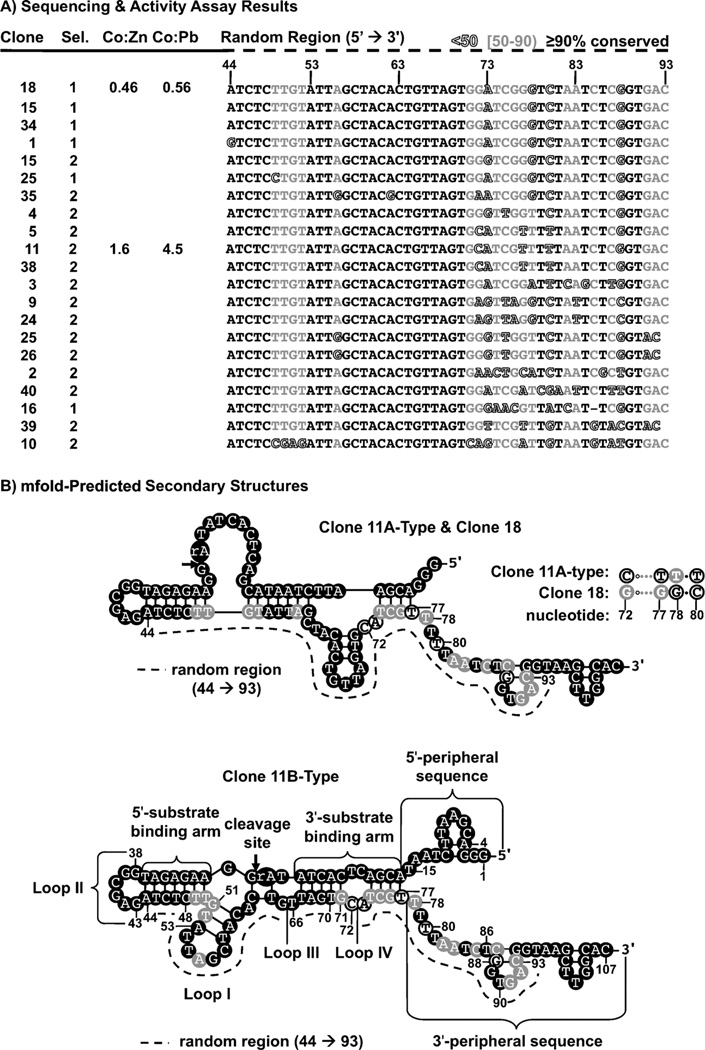
Figure 1.
The artificial phylogenetic analysis and secondary structures of the clone 11 and clone 18 DNAzymes. A) The aligned sequences from selections 1 and 2 that were used for artificial phylogenetic analysis. Sequences are arranged according to similarity. Shading indicates the degree of nucleotide conservation: positions that were highly (>90%) conserved are shown in black, positions that were moderately (50–90%) conserved are shown in grey, and nonconserved (<50%) positions are outlined. Positions 71–83 were highly variable. Nucleotide positions are based on the full-length clone 11 sequence. B) The sequence variability of each sequence overlaid with their mfold-predicted secondary structures. Cleavage sites are indicated by arrows. Two secondary structures were predicted for clone 11: the 11A-type and the 11B-type secondary structure. Clone 18 differs from clone 11 by only four nucleotides (72, 77, 78 and 80), and the single secondary structure predicted for clone 18 is identical to that of the 11A-type secondary structure.