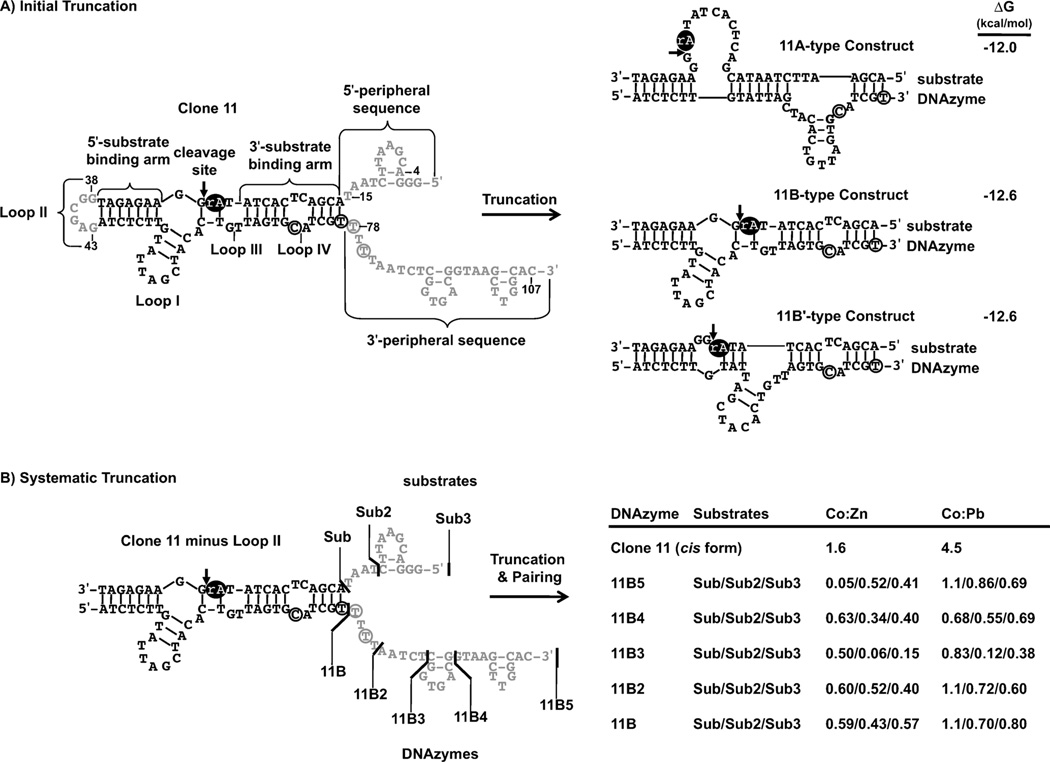
Figure 2.
Truncation of the clone 11 cis-cleaving DNAzyme. The portions of clone 11 that were removed are shown in grey and the ribonucleotide is shown as a black oval. A) The clone 11 cis-cleaving DNAzyme was initially truncated to the 11B-trans cleaving DNAzyme (11B-trans) by deleting loop II and all peripheral sequences. Mfold predicted three different secondary structures for 11B-trans: 11A-type, 11B-type, and 11B’ (shown here with corresponding ΔG values). The 11B and 11B’ structures differ only at positions 51–67, and 11B’ has a single-stranded region immediately following the cleavage site. Only 5–6 base pairs differ between 11B and 11B’. B) The clone 11 cis-cleaving DNAzyme was then modified by deleting loop II and systematically truncating peripheral sequences by small increments. This produced three substrate variants (Sub3, Sub2 and Sub) and five DNAzyme variants (11B5, 11B4, 11B3, 11B2 and 11B). These sequences were paired in every possible permutation and their metal-dependent activities were measured. Co:Zn and Co:Pb selectivity indices for each permutation are reported for the results using Co2+, Zn2+, and Pb2+ (500 µm). The full results are listed in Table 2.