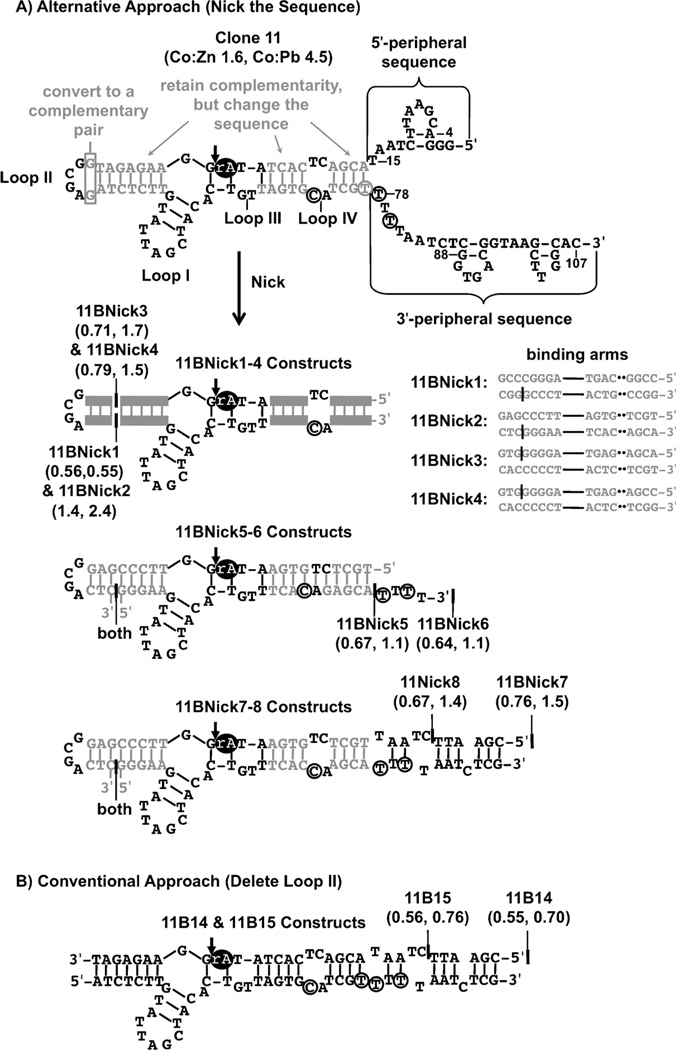
Figure 3.
Truncation of the clone 11 cis-cleaving DNAzyme by conventional and alternative truncation approaches. The portions of clone 11 that were changed during truncation are shown in grey. The ribonucleotide is shown as a black oval. A) An alternative truncation approach to the conventional method (shown in Figure 2B) preserved the secondary structure of 11B, retained many conserved nucleotides from clone 11, and preserved the substrate binding arm base pairs. The nucleotides within clone 11’s binding arms were modified, but its base pairing interactions were preserved. Then the 5’ substrate-binding arm was nicked at one of two positions. Thus the two resulting truncated constructs retained loop II. In order to preserve the base pairing in the 5’-substrate binding arm, two nucleotides in loop II were base-paired, converting loop II to a tetraloop. The peripheral sequences were then systematically truncated. In 11BNick5 and 11BNick6, two additional point mutations were introduced in loop IV to increase the substrate binding arm complementarity. The mfold-predicted secondary structures of each of the constructs obtained by the alternative approach (11BNick1–8) are shown. 11BNick1–4 have secondary structures that differ only in the substrate binding arm sequences, as shown. Their Co:Zn and Co:Pb selectivity indices are shown in parentheses. B) The truncation of clone 11 by the conventional approach described in Figure 2B produced two additional constructs: 11B14 and 11B15. Modifications included 1) deleting loop II and 2) systematically truncating the peripheral sequences.