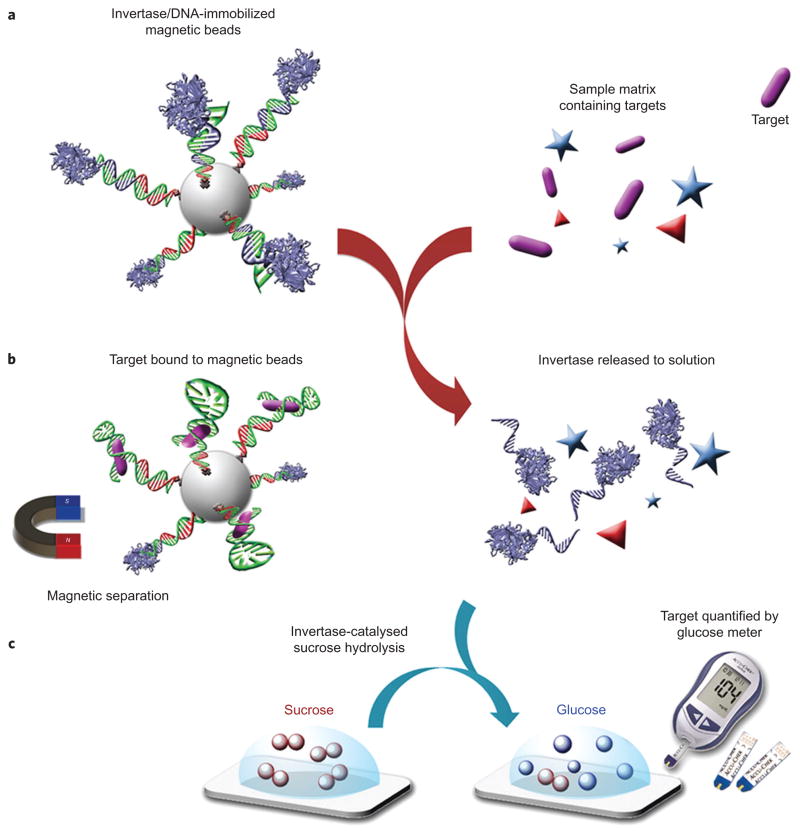
Figure 1.
Schematic of method using a PGM to detect a wide range of targets beyond glucose. a, DNA–invertase conjugates are immobilized onto magnetic beads (MBs) by DNA hybridization with a functional DNA that can specifically respond to the target of interest. b, Following addition of the target, interaction between the functional DNA and its target perturbs the DNA hybridization and causes the release of DNA–invertase conjugates from MBs into solution. c, After removal of MBs by a magnet, the DNA–invertase conjugates in solution can efficiently catalyse the hydrolysis of sucrose into glucose, which is quantified by a glucose meter. Because the concentration of DNA–invertase conjugate released into solution is proportional to the concentration of target present in the sample, the readout in the PGM can be used to quantify the target concentration. (More than one DNA strand is conjugated to each invertase, but the scheme here shows only one DNA strand on each invertase for reasons of clarity.).