Abstract
The nematode Caenorhabditis elegans uses striated muscle in its body wall for locomotion. The myofilament lattice is organized such that all the thin filament attachment structures (dense bodies, analogous to Z-disks) and thick filament organizing centers (M-lines) are attached to the muscle cell membrane. Thus, the force of muscle contraction is transmitted through these structures and allows locomotion of the worm. Dense bodies and M-lines are compositionally similar to focal adhesions and costameres, and are based on integrin and associated proteins. Null mutants for many of the newly discovered dense body and M-line proteins do not have obvious locomotion defects when observed casually, or when assayed by counting the number of times a worm moves back and forth in liquid. We hypothesized that many of these proteins, located as they are in muscle focal adhesions, function in force transmission, but we had not used an appropriate or sufficiently sensitive assay to reveal this function. Recently, we have developed a new quantitative assay of C. elegans locomotion that measures the maximum bending amplitude of an adult worm as it moves backwards. The assay had been used to reveal locomotion defects for null mutants of genes encoding ATN-1 (α-actinin) and PKN-1 (protein kinase N). Here, we describe the details of this method, and apply it to 21 loss of function mutants in 17 additional genes, most of which encode components of muscle attachment structures. As compared to wild type, mutations in 11 genes were found to have less ability to bend, and mutations in one gene were found to have greater ability to bend. Loss of function mutants for eight proteins had been reported to have normal locomotion (ZYX-1(zyxin), ALP-1 (Enigma), DIM-1, SCPL-1), or locomotion that was not previously investigated (FRG-1 (FRG1), KIN-32 (focal adhesion kinase, LIM-8), or had only slightly decreased locomotion (PFN-3 (profilin)).
Keywords: muscle, sarcomere, muscle focal adhesions, C. elegans, muscle genes, locomotion assay
INTRODUCTION
The sarcomere is the fundamental unit of muscle contraction. Hundreds of proteins need to be precisely assembled into this highly organized structure. New components of the sarcomere are discovered each year. Despite this cataloging, we have a poor understanding of the assembly, maintenance, and function of most of these proteins.
The nematode C. elegans is an excellent system for investigating muscle biology. This is due to its facile (forward and reverse) genetics, availability of many mutants, and transparent body (which enables evaluation of muscle structure by polarized light and localization of green fluorescent protein (GFP) tagged proteins)[1–4]. Many components of the C. elegans sarcomere and its membrane-extracellular matrix (ECM) attachment structures have been defined and most were first identified through mutations. Most of the mutants fall into one of two phenotypic classes: the “Unc” class (uncoordinated movement) comprising 40 genes [5,6], and the “Pat” class (paralyzed arrested at two-fold stage in embryonic development) comprising 20 genes [7,8].
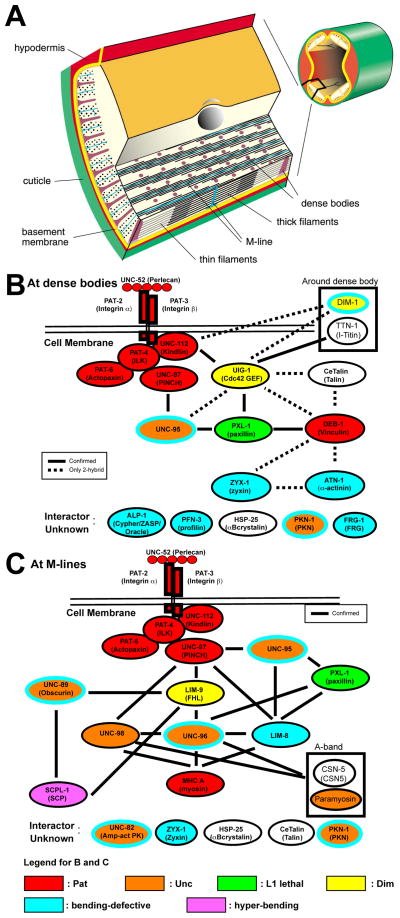
In C. elegans, the body wall muscle is essential for its locomotion. The sarcomeric region is restricted to a narrow ~1.5 μm zone adjacent to the cell membrane along the outer side of the muscle cells [1] (Fig. 1A). All of the Z-disk-like structures (called dense bodies) and M-lines are anchored to the muscle cell membrane via integrin containing focal adhesions [2–4]. In this way, the force of muscle contraction is transmitted directly through the muscle cell membrane, the ECM, overlying hypodermis and cuticle to outside the worm. Over the past 8 years, it has been reported that multiple protein complexes link the ECM to thick filaments at the M-line in C. elegans (Fig. 1C): Concentrated at the base of M-lines is UNC-52 (perlecan), where it presumably interacts with the extracellular portions of PAT-2 (α-integrin) and PAT-3 (β-integrin) embedded in the muscle cell membrane. The cytoplasmic tail of PAT-3 (β-integrin) is associated with a complex of four conserved proteins (UNC-112 (Kindlin), PAT-4 (integrin linked kinase, ILK), PAT-6 (Actopaxin) and UNC-97 (PINCH)) [9–11]. UNC-97 links to myosin in thick filaments through four different complexes: via UNC-98, via LIM-9 (FHL) and UNC-96, via LIM-8, and via UNC-95 and LIM-8 [12–14]. Similar progress is being made in defining a dense body protein interaction matrix that explains linkage of the muscle cell membrane to thin filaments (Fig. 1B) [15]. Like the M-line, at the dense body, the integrin tail is associated with the UNC-112/PAT-4/PAT-6/UNC-97 four-protein complex. In addition, there is a three-protein complex consisting of CeTalin/DEB-1 (vinculin)/ATN-1 (α-actinin) that is specific for dense bodies, and Z-disks in other animals. An interesting feature of what we know about the dense body interacting proteins is that there is a molecular linkage between the UNC-112 four-protein complex, and the CeTalin three-protein complex. The linkage consists of the intermediary proteins, UIG-1 (a Cdc42 GEF) and PXL-1 (paxillin)(Fig. 1B). Such a linkage between these two protein complexes has not been described before in any other system. Finally, although for the nematode dense body, the molecular linkage to thin filaments has not yet been clarified, it is most likely through ATN-1 (α-actinin) and DEB-1 (vinculin), as these are well-known F-actin binding proteins in other systems.
Figure 1.
Muscle structure and sets of interacting proteins found at muscle focal adhesions (M-lines and dense bodies) in C. elegans striated muscle. (A) Organization of the myofilament lattice in body wall muscle of C. elegans. The upper right depicts a portion of a nematode cut in cross section with body wall muscle quadrants in yellow. The enlargement shows the organization of filaments and membrane attachment structures in several planes of section. The sarcomeric region is confined to a narrow zone adjacent to the cell membrane along the outer side of the muscle cells. Note that all the dense bodies (Z-disk analogs) and M-lines are anchored to the muscle cell membrane and thus positioned to transmit the force of muscle contraction. (B and C) Sets of interacting proteins identified at dense bodies and M-lines that begin with UNC-52 (perlecan) in the ECM and integrins in the muscle cell membrane, and continue until the thin and thick filaments, respectively. Vertebrate homologs or orthologs are indicated in parentheses. The references for the dense body interactions are in reference [15], and unpublished data from the Benian and Moerman labs; the references for the M-line interactions are in references [4] and [15], and unpublished from the Benian lab. As shown at the bottom of the figure, the diagrams are color-coded to reflect loss of function mutant phenotype: red, Pat embryonic lethal; orange, Unc adults; yellow, Dim (disorganized myofibrils); green, L1 larval lethal; yellow, Dim (disorganized muscle); blue, bending-defective; purple, hyper-bending. The bending-defective (blue) and hyper-bending (purple) phenotypes were identified using the new locomotion assay described here. Note that mutants for five proteins, UNC-95, PKN-1, UNC-89, UNC-96, and UNC-82, are both Unc (orange) and bending defective (blue outer oval). DIM-1 is both Dim (yellow) and bending defective (blue outer oval). In addition to those proteins with Pat or L1 lethal phenotypes, bending assays were not conducted for TTN-1, CeTalin, paramyosin or CSN-5. HSP-25 was tested, but did not show a bending defect.
Null mutants for many of these newly discovered M-line and dense body proteins do not have locomotion defects when observed casually or when assessed by a conventional motility assay in which the number of times a worm moves back and forth in liquid are counted [16]. We hypothesize that these proteins, located as they are at muscle focal adhesions function in force transmission, but an appropriate assay was not used to reveal their functions. Recently, we have applied a new, more quantitative assay of nematode locomotion and used it to define a motility defect for a mutant that had previously been thought not to have one. This assay measures the maximum amount of bending the adult worm makes as it moves backwards upon anterior mechanical stimulation. Our test case for the assay was the null mutant of the single α-actinin gene in the nematode, atn-1 [17]. atn-1(ok84) null mutants have abnormally short and broad dense bodies and yet display normal movement on an agar surface by casual observation, and normal swimming in liquid. In contrast, our assay showed that this mutant has a reduced ability to bend as compared to wild type [17]. The bending aspect of locomotion was restored to wild type levels in mutant animals carrying integrated copies of the wild type atn-1 gene. Thus, the loss of α-actinin, a major component of dense bodies, results in nematodes that are less able to bend, perhaps because they transmit the force generated by myosin/actin interaction, less efficiently. This reduced ability to bend is not specific to loss of function of α-actinin, as we have recently reported that mutations in pkn-1 also show this phenotype [18]. pkn-1 encodes the nematode ortholog of protein kinase N (PKN). Either loss of function by RNAi or genomic deletion, or overexpression of the kinase domain of PKN-1 in muscle, results in a “loopy Unc phenotype”, in addition to reduced maximal bending [18]. A GFP fusion of PKN-1 (residues 1–138) is localized to dense bodies and M-lines. Vertebrate PKN is known to interact with α-actinin [19].
Curiously, when we measured the maximum wave amplitude as adult worms moved forwards, we could not discern a difference between wild type and atn-1(ok84)(Gina Cremona, J. Stirman and H. Lu, unpub. data). We hypothesize that the worm may exert more force and power when moving backwards during an escape behavior (e.g. upon anterior touch) than moving forwards (e.g. during foraging). If so, there may be both an anatomical and ecological/evolutionary explanation. The body wall muscles are organized into 4 quadrants containing two rows of spindle-shaped cells in each quadrant. In the front half of the animal, the cells in each quadrant are arranged almost in pairs, whereas in the back half of the animal, the cells are organized in a more alternating fashion. Therefore, two thirds of all the muscle cells are located anterior to the vulva [20]. Initiation of backward movement begins with a wave of contraction in the anterior half of the animal; therefore, more force may be generated during backward movement. Another possibility is that C. elegans has been selected to exert its maximum initial force and speed in the reverse direction to escape from predatory fungi that use inflatable constricting rings to trap nematodes [21].
In this paper, we describe more fully the new assay and apply it to 21 loss of function mutants in 17 additional muscle genes, many of which had no previously reported motility defects. These genes encode proteins that are known, or are suspected to be, localized to muscle attachment structures (dense bodies and M-lines). Mutations in 11 of these genes were found to have less than wild type maximum bending amplitude when in reversal. Mutations in one gene, were found, remarkably, to have a greater than wild type ability to bend.
MATERIALS AND METHODS
C. elegans strains
Nematodes were obtained from the Caenorhabditis Genetics Center and from the Mitani lab at Tokyo Women’s Medical University School of Medicine. N2 (Bristol) was the wild type strain. The following strains had been outcrossed to wild type to remove background mutations: dim-1(ra102), unc-82(e1323), unc-95(su33), atn-1(ok84), lim-8(ok941), unc-98(sf19), unc-96(sf18), zyx-1(gk190), unc-89(tm752), and unc-89(su75). Although the scpl-1 and lim-9 mutants had not been outcrossed, similar bending assay results were obtained for two independently isolated alleles of each gene (hyper-bending for scpl-1(gk283) and scpl-1(ok1080), and hypo-bending for lim-9(gk210) and lim-9(gk106)).
Assay Preparation
All strains of C. elegans were grown at 20° C using a standard protocol on NGM (Nematode Growth Medium) agar plates seeded with E. coli strain OP50 [22]. Plates used for the assay were 2.1% NGM plates without bacteria growing on them. The plates were allowed to dry for at least 48 hours in a laminar hood after pouring and up to two weeks after date of pouring. When assay plates were not being used they were stored upside-down at 4°C. Plates were allowed to adjust to room temperature for at least 2 hours before being used.
Twenty young adult worms of each mutant strain were assayed together with 5 N2 (wildtype) worms each time the assay was conducted. N2 was tested along with each mutant strain on the same lot of plates and at the same time to ensure the plate’s age, the humidity, slight variance in temperature, the plate’s moisture, and other various factors are controlled for. No less than five worms of a specific strain were picked by an individual not performing the assay, and transferred to an assay plate labeled with a random integer 1–5. This was repeated three more times with each plate being labeled a different integer. No less than five N2 worms were picked in the same manner onto a plate labeled with the remaining number 1–5. The plate numbers and corresponding strain were recorded. The strain being tested, and which plate contained wild-type, were not revealed to the individual performing the assay to ensure the individual would not bias the results.
The Assay
A ZEISS Stemi SV 6 dissecting microscope and a THORLABS DCC1545M microscope camera were used to record the worm’s movement on an assay plate. The camera was attached to the microscope via a 0.5x c-mount. Videos were recorded using the THORLABS video acquisition program (uc480 viewer). The illumination was adjusted to provide maximum contrast between the worms and the assay plate. Worms were picked individually onto an assay plate and allowed to crawl freely for 2 minutes. After this time, the worm was tapped gently on the head to induce reverse movement and the behavioral response was recorded. To keep the worm in the field of view, the assay plate was gently moved with the worm’s movement. The video capture was halted once the worm began to move forward again. The worm was then allowed to crawl freely for one minute, and was then tapped and recorded again in the same manner as before. This was repeated about five times for each worm, each one minute apart. This entire process was repeated for each worm in the population being assayed.
Analysis
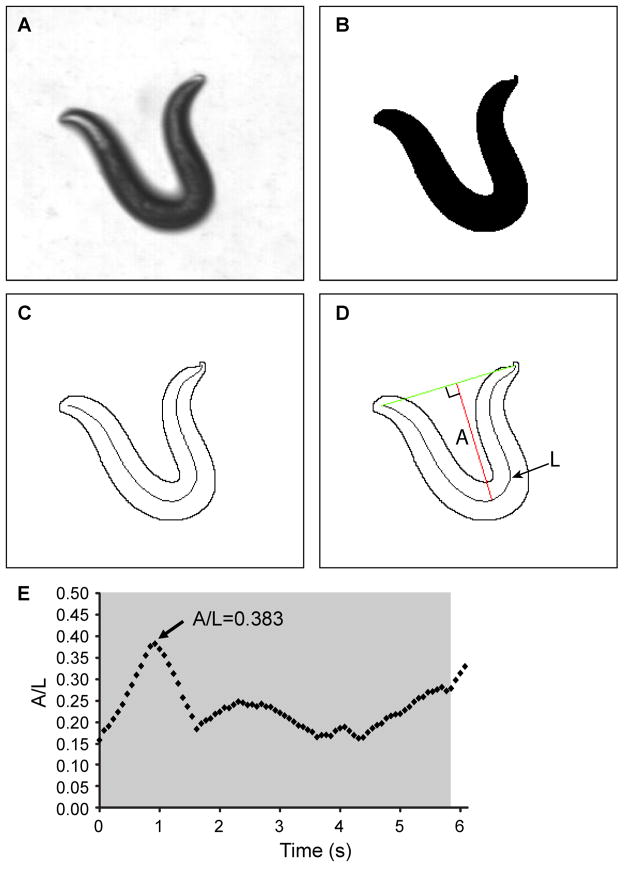
The videos were analyzed using a custom-written LabVIEW program that thresholds each individual frame of the video to identify the worm from the background. A line (spline) was then automatically fitted to the mid-body of the worm along the entire length of its body [23], and we define the length of this line as “L” (Fig. 2D). The amplitude, “A”, of the curve of the worm’s body was then calculated using the spline and was defined as the maximum perpendicular distance from the spline to a line connecting the tip of the head to the tail. To account for the differing body lengths of the worms, the ratio of the bend amplitude to body length (“A/L”) was calculated and recorded as the primary measurement for the worm’s ability to bend. The maximum A/L value for each video clip of the worm moving backwards was recorded and averaged with the same worm’s A/L value from the other video clips. Maximum A/L values for all individual worms of the same genetic background were averaged and reported.
Figure 2.
Measurement of the amplitude to length ratio in a reversal. (A) Frame from video showing deep bend of an N2 nematode in a reversal. (B) Binary image of the frame from (A). (C) Outline of the animal and the corresponding midline spline used to determine the length of the animal. (D) Measurement of the amplitude of the bend was made by measuring the longest perpendicular length, A, from a line connecting the head to the tail to the midline spline, of contour length L. The ratio of this value, A, to the contour length, L, was calculated (A/L). (E) Time course measurement of the A/L values for a single measurement. The tap stimulus was applied shortly before t=0 seconds (s). The grey region indicates the time the animal is in a reversal and it is during this time that the maximum A/L value is measured.
Statistical Analysis
A Kolmogorov–Smirnov normality test was conducted on all of the mutants individually to determine if the data were distributed normally. All of the data were then tested with a Kruskal-Wallis one-way analysis of variance to determine if the mutants showed any difference between samples. A Kruskal-Wallis one-way analysis of variance was used again to determine if the N2 controls showed variation day-to-day. A Mann-Whitney U test was used to perform pairwise comparisons between each mutant and its respective day-to-day N2 control. The p-values resulting from this test are shown in Table 1 in the column, “p-values when tested w/day-to-day controls”. A Mann-Whitney U test was also used to perform pairwise comparisons between each mutant and all of the N2 maximum amplitude to length ratios pooled together (n=130). The p-values resulting from this test are shown in Table 1 in the column, “p-values when tested w/all N2”.
Table 1.
Mutants Ability to Bend
| Mutant | Mean of Maximum A/L Ratios | Std. Error | N2 Day-to-Day-Control Mean | Std. Error | p-values when tested w/ N2 Day-to-Day Controls | p-values when tested w/ all N2 | ||
|---|---|---|---|---|---|---|---|---|
| scpl-1 (gk283) | 0.37092 | 0.00523 | 0.32290 | 0.01150 | 0.000 | †† | 0.000 | †† |
| scpl-1 (ok1080) | 0.36697 | 0.00434 | 0.34040 | 0.01030 | 0.016 | † | 0.000 | †† |
| unc-98 (sf19) | 0.34878 | 0.00666 | 0.36950 | 0.01020 | 0.117 | N/S | 0.158 | N/S |
| lim-9 (gk210) | 0.34575 | 0.00547 | 0.36390 | 0.01180 | 0.162 | N/S | 0.867 | N/S |
| *lim-8 (ok941) | 0.34367 | 0.00594 | 0.39049 | 0.00579 | 0.000 | †† | 0.799 | N/S |
| uig-1 (ok884) | 0.33981 | 0.00616 | 0.34980 | 0.01090 | 0.340 | N/S | 0.605 | N/S |
| hsp-25 (tm700) | 0.33951 | 0.00652 | 0.35134 | 0.00758 | 0.778 | N/S | 0.844 | N/S |
| atn-1 Rescue | 0.33928 | 0.00768 | 0.36639 | 0.00934 | 0.131 | N/S | 0.820 | N/S |
| lim-9 (gk106) | 0.33904 | 0.00556 | 0.34430 | 0.01550 | 0.481 | N/S | 0.387 | N/S |
| tag-163 (ok644) | 0.33624 | 0.00576 | 0.34920 | 0.01220 | 0.227 | N/S | 0.148 | N/S |
| *unc-96 (sf18) | 0.33143 | 0.00592 | 0.40391 | 0.00291 | 0.000 | †† | 0.094 | N/S |
| kin-32 (ok166) | 0.32132 | 0.00636 | 0.34980 | 0.01400 | 0.023 | † | 0.007 | †† |
| **pfn-3(tm1362) | 0.32073 | 0.00757 | 0.34050 | 0.01520 | 0.196 | N/S | 0.032 | † |
| alp-1(tm1137) | 0.31293 | 0.00683 | 0.37360 | 0.00799 | 0.000 | †† | 0.000 | †† |
| pkn-1 (ok2273) | 0.30609 | 0.00804 | 0.35376 | 0.00970 | 0.008 | †† | 0.000 | †† |
| atn-1(ok84) | 0.30583 | 0.00718 | 0.36639 | 0.00934 | 0.000 | †† | 0.000 | †† |
| zyx-1 (gk190) | 0.30439 | 0.00602 | 0.33330 | 0.01300 | 0.015 | † | 0.000 | †† |
| unc-89 (su75) | 0.29632 | 0.00633 | - | - | - | N/A | 0.000 | †† |
| dim-1(ra102) | 0.29309 | 0.00648 | 0.34540 | 0.01170 | 0.001 | †† | 0.000 | †† |
| unc-89 (tm752) | 0.29098 | 0.00765 | 0.34710 | 0.01320 | 0.002 | †† | 0.000 | †† |
| frg-1 (tm2803) | 0.27581 | 0.00785 | - | - | - | N/A | 0.000 | †† |
| unc-95(su33) | 0.22771 | 0.00717 | 0.34660 | 0.01350 | 0.000 | †† | 0.000 | †† |
| unc-95(ok893) | 0.19587 | 0.00864 | 0.34660 | 0.01350 | 0.000 | †† | 0.000 | †† |
| unc-82(e1323) | 0.18248 | 0.00608 | 0.34250 | 0.01120 | 0.000 | †† | 0.000 | †† |
| N2 | 0.34587 | 0.00352 | N/A | N/A | N/A | N/A | ||
N/S indicates “Not Significant” and p>0.05.
indicates 0.05>p>0.01.
indicates p<0.01.
A “ * ” by the mutant genotype indicates the mutant was shown to be statistically different from N2 when analyzed with the N2 on the same day (“day-to-day controls”) but statistically the same as N2 when analyzed with all N2.
A “ ** ” by the mutant genotype indicates the mutant was shown to be statistically the same as N2 when analyzed with the N2 on the same day (“day-to-day controls”) but statistically different from N2 when analyzed with all N2. Mutants where both analyses agreed were left un-starred.
unc-89 (su75) and frg-1 (tm2803) were not tested with an N2 day-to-day control.
RESULTS AND DISCUSSION
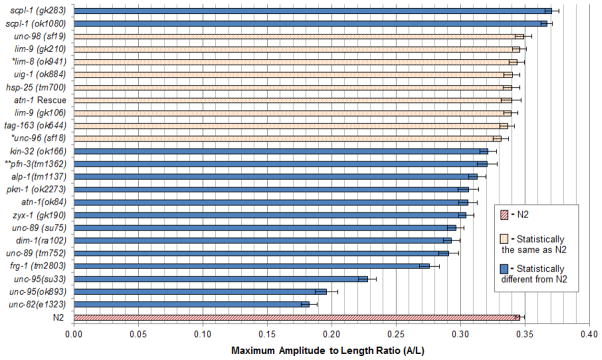
A Kolmogorov–Smirnov normality test indicated that the data were not distributed normally with a high degree of certainty (p<0.05 for all samples). For this reason, non-parametric statistical analyses were used. A Kruskal-Wallis one-way analysis of variance showed there was variation amongst N2 controls day-to-day with a p-value of 0.000. Because of this, a Mann-Whitney U test was used to make pairwise comparisons between mutants and their day-to-day control to account for this variation. The p-values for these pairwise comparisons are recorded in Table 1 in the column labeled “p-values when tested w/day-to-day controls”. A Mann-Whitney U test was also used to compare each mutant to all of the N2 maximum amplitude to length ratios pooled together. The p-values for these pairwise comparisons are recorded in Table 1 in the column labeled “p-values when tested w/all N2”. Except for three mutants (pfn-3, unc-96, and lim-8), the results from the two methods of comparison, agree. Fig. 3 shows the average maximum amplitude to length ratios for each mutant, for all of N2, and whether these values from each mutant were statistically different from all of N2.
Figure 3.
Graphical display of the average maximum amplitude to length ratios for all mutants tested and for N2 (wild type). For each mutant, n=20. Error bars indicate standard error of the mean. The legend and coloration refer only to p-values determined from pairwise comparisons with all measurements of N2. More detailed results are displayed in Table 1. *: statistically different from N2 when analyzed with N2 on the same day (“day-to-day control”). **: statistically the same as N2 when analyzed with N2 on the same day (“day-to-day control”).
Mutations in 11 of 17 genes, in addition to the “positive controls” atn-1 and pkn-1, were found to have less than wild type maximum bending amplitude (Fig. 3). Of these 11 genes, four of them, unc-82, unc-95, unc-89 and unc-96 were previously described as having reduced motility. unc-82 mutants were originally isolated during a screen for mutants having a disorganized myofilament lattice by polarized light microscopy, and described as moving “perceptibly slower” than wild type animals [5]. Thus, our observed defect in bending of the null allele, unc-82(e1323), is not unexpected. unc-82 mutants display defects in localization of thick filament and M-line components [24]. unc-82 encodes a set of polypeptides as large as 1793 aa, with a protein kinase domain near its N-terminus, and is related to ARK5/SNARK, PAR-1, and SNF1/AMP-activated protein kinase families [24]. UNC-82::GFP is localized to the M-lines [24].
unc-95 mutants were originally isolated in a motility-requiring selection, and were described as having disorganized sarcomeres by polarized light and electron microscopy, and as being slow to paralyzed in movement [6]. Thus, as with unc-82 mutants, our observed defect in bending of unc-95 mutants (detected in two mutant alleles, one of which is an intragenic deletion), is not unexpected. UNC-95 is a novel protein with a single LIM domain at its C-terminus [25], and is localized to M-lines, dense bodies, and muscle cell nuclei by a GFP fusion [25], and M-lines and dense bodies by antibodies [14].
Our assay also demonstrated a bending defect for two mutant alleles of unc-89. unc-89 encodes a giant polypeptide located at the M-line [26–28], homologous to human obscurin. In unc-89 mutants, by polarized light microscopy, the body wall muscle sarcomeres are disorganized [5,27], with A-bands forming a “basket-weave” pattern. By electron microscopy, unc-89 mutants show a thinner sarcomeric region and usually lack M-lines [5,29]. Despite the disruption in sarcomeric structure, casual observation of worms moving on a plate cannot reliably distinguish unc-89 mutants from wild type. Nevertheless, using a standard swimming assay, unc-89 mutants are slower than wild type (T. Tinley, T. Ferrara, G. Benian, unpub. data). Thus, the observed reduction in maximal bending of unc-89 mutants is also consistent with the previously known reduction in swimming ability of these mutants.
The original allele of unc-96, su151, was isolated in the same motility-requiring selection, as was performed to isolate a mutant in unc-95 [6]. By polarized light microscopy, unc-96 mutants display reduced myofibrillar organization and characteristic birefringent “needles” at the ends of the muscle cell [6, 30]. unc-96 mutants show major defects in the organization of M-lines, and in the localization of a major invertebrate-specific thick filament protein, paramyosin [30]. unc-96 encodes a novel protein that is localized to M-lines and interacts with UNC-98, paramyosin and CSN-5, a component of the COP9 signalosome complex [30, 31]. By the thrashing assay, the likely null allele of unc-96, sf18, displays an approx. 40% reduction in motility as compared to wild type [30]. Thus, it was surprising that the bending assay did not reveal a defect in bending, when the result was compared to the average of all wild type results (Fig. 3). Nevertheless, when bending on sf18 was compared to bending of wild type animals conducted on the same day and same plates, a statistically significant difference was observed (Table 1).
Therefore, our new assay can detect a locomotion defect in mutants (unc-82, unc-95, unc-89, and possibly unc-96) that are well-recognized to have an adult “Unc” phenotype. It was thus unexpected for us to find that the maximal bending ability of unc-98(sf19) was not different from wild type. This is in contrast to the fact that by swimming assay, unc-98(sf19)[12] is significantly slower than wild type (30–40% reduction). Therefore, results with unc-98 and possibly unc-96 mutants suggest that our new bending assay and the swimming (thrashing) assay measure different aspects of nematode motility.
Remarkably, loss of function of one of the genes, scpl-1, represented by 2 mutant alleles (gk283 and ok1080), showed increased maximal bending ability. scpl-1 encodes a CTD-type phosphatase localized to sarcomeric M-lines and I-bands, and was isolated as a binding partner for the protein kinase domains of UNC-89 [32]. It is certainly intriguing that loss-of-function for these binding partners, UNC-89 and SCPL-1, have opposite phenotypes. gk283 and ok1080 do not show any obvious defects in muscle structure, or in motility in a swimming assay (our unpublished data). We can only speculate as to the mechanism by which the scpl-1 mutants are hyper-bending. We know that in order for the worm to achieve alternating sinusoidal locomotion, when the worm bends, the muscles within the bend contract at the same time as muscles on the opposite side of the worm relax. Therefore, the bending amplitude is a sum of these two activities. Thus, the hyper-bending of scpl-1 mutants might result from either increased contraction of the bending muscles, or increased relaxation of the opposite muscles. The fact that unc-89 and scpl-1 mutants have opposite phenotypes, and unc-89 mutants, with their disorganized myofibrils are less able to generate or transmit force, suggests that hyper-bending of scpl-1 is due to increased relaxation.
Mutants in the zyx-1, frg-1, alp-1, kin-32, pfn-3, and lim-8 genes, were also found to be bending-defective. These genes encode, respectively, zyxin, FRG-1 (ortholog of a candidate protein involved in human FSH muscular dystrophy), ALP-1 (Enigma homolog), KIN-32 (focal adhesion kinase, FAK), PFN-3 (one of three profilins), and LIM-8 (a LIM domain protein). All of these proteins are known or suspected to be located at dense bodies or I-bands, and most are known or suspected to function with α-actinin. α-actinin, a well studied F-actin crosslinking protein, is a major component of nematode dense bodies and vertebrate muscle Z disks [33,17].
A ZYX-1-GFP fusion has been shown to localize to dense bodies, M-lines and muscle cell nuclei [34]; ZYX-1 interacts with ATN-1 via a 2-hybrid assay (H. Qadota and G. Benian, pers. comm.). Antibodies to FRG-1 localize to dense bodies, and FRG-1 is mis-localized in atn-1(ok84)[35]. ALP-1-GFP fusion proteins are localized to dense bodies, cell-cell junctions and muscle cell nuclei [36]. Antibodies to ALP-1 localize to dense bodies, co-localizing with α-actinin, but not vinculin; in atn-1(ok84), ALP-1 is not restricted to dense bodies. A putative null intragenic deletion allele of zyx-1, zyx-1(gk190) was reported to have “no obvious behavioral phenotype” [34]. A loss-of-function mutant for frg-1 was not described in the study on the frg-1 gene by Liu et al. (2010)[35], probably because it was not previously available. A null allele for alp-1 displays a normal swimming assay (liquid motility assay)[37]. A 50% reduction in atn-1 activity (atn-1/+) resulted in enhancement of the actin aggregation phenotype of an alp-1 mutant [37]. Although a direct interaction between nematode ALP-1 and ATN-1 has not been reported, the vertebrate ALP-1 homologs have been shown to interact with α-actinin (e.g [38]). KIN-32 is the nematode ortholog of focal adhesion kinase, and although the nematode protein has not yet been localized, it is likely to be located at dense bodies and/or M-lines, given its well known localization to focal adhesions in vertebrate cells [39]. pfn-3 encodes one of three profilins in C. elegans, and pfn-3 is specifically expressed in body wall muscle cells [40]. Anti-PFN-3 antibodies localize to the cytoplasmic tips of dense bodies, partially co-localizing with α-actinin, but PFN-3 extends more distally (deeper into the cytoplasm). A swimming assay of an intragenic deletion allele, pfn-3(tm1362), which shows no detectable PFN-3 by western blot, was “slightly slower” than wild type, although tests for statistical significance were not shown [40]. Nevertheless, our data are consistent with this report, and indeed pfn-3(tm1362) displays a statistically significant reduction in maximal bending ability (Fig. 3). It should be noted, however, that when the bending assay results for pfn-3(tm1362) were compared to wild type tested on the same day, no difference was noted (Table 1).
LIM-8, contains one LIM domain and one PDZ domain, and was identified as a binding partner for UNC-97 (PINCH) [14]. Antibodies localize LIM-8 to M-lines and I-bands. At M-lines, LIM-8 also interacts with UNC-95, UNC-96, myosin (MHC A) [14], and with paxillin (PXL-1) [15]. Although a null allele lim-8(ok941) has no defect by casual observation or by swimming assay (H. Qadota and G. Benian, unpub. data), it did show a defect in bending, but only when results were compared to wild type assayed on the same day and plate (compare Table 1 and Fig. 3). Nonetheless, the functional importance of lim-8 for body wall muscle is also suggested by the fact that although lim-8(ok941) has normal organization of myofilaments, mild disorganization is seen when RNAi for lim-8 was conducted on worms in which body wall muscle was deficient in pxl-1 [15].
Given that loss of function for atn-1 results in reduced maximal bending [17], the fact that five suspected α-actinin interacting molecules also show defective bending suggests that each of these molecules works together with α-actinin in transmitting force at dense bodies.
Our results on the dim-1 mutant are quite interesting. dim-1 encodes a novel polypeptide containing 3 Ig domains, and GFP tagged DIM-1 localizes around and between dense bodies [41]. Multiple loss of function alleles of dim-1 were isolated as extragenic suppressors of an unc-112 hypomorphic Unc allele, which by itself displays disorganized myofibrils and near paralysis as an adult [41]. The only phenotype reported by Rogalski et al. (2003)[41] for dim-1 mutant alleles, all of which a null alleles, was a slight disorganization of the myofilament lattice when viewed by polarized light. In their report, no mention was made as to whether such mutants were obviously slow moving by direct observation, or by swimming assay. (Indeed, we have observed that dim-1(ra102) behaves like wild type, as it moves on an agar plate.) Therefore, our finding that dim-1(ra102) has reduced maximal bending compared to wild type indicates a new function for dim-1 in nematode locomotion. It should be pointed out that Rogalski et al. (2003)[41], because they did not observe a motility defect for dim-1 mutants, in contrast to typical Unc mutants which display both disorganized myofibrils and reduced motility, created a new phenotypic category of muscle genes, dim, for disorganized muscle.
Among the proteins that are components of M-lines and dense bodies (Fig. 1), loss of function mutants for uig-1 [42] and lim-9 [14,8], also would be considered members of the Dim category (colored yellow in Fig. 1). In contrast to dim-1, our bending assay on two null alleles of lim-9 and one null allele of uig-1, did not reveal a bending defect. UIG-1 is a Cdc42 specific guanine nucleotide exchange factor located at dense bodies [42], and LIM-9 is the nematode protein most homologous to human FHL (four and a half LIM domains protein) and is located at M-lines and I-bands [14, 8]. Thus, based on our assays, these two proteins have no crucial role in motility, or alternatively, we have not yet employed the proper assay to reveal a motility function. In addition, genetic redundancy is suggested as a possibility by our protein interaction matrices (Fig. 1): uig-1 may be redundant with pxl-1 at dense bodies, and lim-9 may be redundant with unc-98 and with lim-8 at M-lines.
Admittedly, we have conducted motility assays on only single alleles for most of these genes. (There are four exceptions —unc-89, lim-9, scpl-1, and unc-95— which are represented by two alleles each.) In order to verify that the reduced bending observed in these mutants is attributable to mutation in any of one these genes, we will need to repeat our assays on additional mutant alleles, and conduct the assays on transgenic animals carrying wild type copies of each gene in question, looking for rescue to wild type bending. Nevertheless, our preliminary results demonstrating that mutations in 2 previously reported (ATN-1 (α-actinin), PKN-1 (PKN)), and 13 new proteins that are components of M-lines, dense bodies, or both structures, have abnormal maximal bending, suggest that this method is valid for detecting the importance of each of these proteins in the locomotion of whole nematodes. Importantly, loss of function mutants for eight of these proteins were either reported to have normal locomotion (ZYX-1 (zyxin), ALP-1 (Enigma), DIM-1, SCPL-1), locomotion that was not previously investigated (FRG-1, KIN-32, LIM-8), or had only slightly decreased locomotion (PFN-3 (profilin)), as compared to wild type nematodes. We expect our method will be useful for analyzing additional muscle attachment genes in the future.
Since the original description of the swimming (thrashing) assay by Epstein and Thomson in 1974 [16], a number of other methods mostly involving video recording and computer/mathematical analysis for quantifying C. elegans motility have appeared in the literature [43–49]. However, most of these methods are designed to analyze complex behaviors such as foraging, chemotaxis and response to vibration, and were not specifically designed or applied to assess muscle mutants. An improvement in determining the swimming (thrashing) frequency that avoids human counting errors, and is rapid and automated and thus applicable for small molecule or mutant screens, was described by Buckingham and Sattelle in 2009 [50]. Nevertheless, as we have demonstrated here and in previous papers [17,18], some muscle mutants do not show a defect in swimming frequency, and yet do show a defect in maximal bending. We contend that our bending assay is a more sensitive measurement of the force generated and/or transmitted by the muscle contractile apparatus. In addition, the swimming and bending assays measure different aspects of nematode locomotion: unc-98 mutants show a swimming defect but have normal maximal bending; atn-1 and alp-1 mutants show bending defects, but not swimming defects.
Acknowledgments
The authors are grateful to Dr. David Garton (Georgia Tech) for his expert advice on statistical analysis. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the NIH. Other strains were obtained from Dr. Shohei Mitani at Tokyo Women’s Medical University School of Medicine. J. F. N. was supported by an REU from the National Science Foundation. G.M.B. thanks the Emory University Research Committee, the Emory University Department of Pathology, and the American Heart Association for financial support. H.L. thanks NIH, NSF and the Sloan Foundation for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waterston RH. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Plainview, New York: 1988. pp. 281–335. [Google Scholar]
- 2.Moerman DG, Fire A. Muscle Structure, Function and Development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C elegans II. Cold Spring Harbor Laboratory Press; Plainview, New York: 1997. pp. 417–470. [PubMed] [Google Scholar]
- 3.Moerman DG, Williams BD. WormBook. The C elegans Research community. 2006. Sarcomere assembly in C. elegans muscle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qadota H, Benian GM. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterston RH, Thomson JN, Brenner S. Devel Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- 6.Zengel JM, Epstein HF. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]
- 7.Williams BD, Waterston RH. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, Khattra J, Rogalski T, Somasiri A, Chaudhry I, Fox RM, Miller DM, Baillie DL, Holt RA, Jones SJ, Marra MA, Moerman DG. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Qadota H, Moerman DG, Williams BD. Curr Biol. 2003;13:922–932. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- 11.Norman KR, Cordes S, Qadota H, Rahmani P, Moerman DG. Devel Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner A, Qadota H, Benian GM, Vogl AW, Moerman DG. Mol Biol Cell. 2011;22:2551–2563. doi: 10.1091/mbc.E10-12-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein HF, Thomson JN. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- 17.Moulder GL, Cremona GH, Duerr J, Stirman JN, Fields SD, Martin W, Qadota H, Benian GM, Lu H, Barstead RJ. J Mol Biol. 2010;403:516–528. doi: 10.1016/j.jmb.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qadota H, Miyauchi T, Nahabedian JF, Stirman JN, Lu H, Amano M, Benian GM, Kaibuchi K. J Mol Biol. 2011;407:222–231. doi: 10.1016/j.jmb.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukai H, Toshimori M, Shibata H, Takanaga H, Kitagawa M, Miyahara M, Shimakawa M, Ono Y. J Biol Chem. 1997;272:4740–4746. doi: 10.1074/jbc.272.8.4740. [DOI] [PubMed] [Google Scholar]
- 20.Hall DH, Altun ZF. C elegans Atlas. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2008. Muscle System; pp. 145–188. [Google Scholar]
- 21.Maguire SM, Clark CM, Nunnari J, Pirri JK, Alkema MJ. Curr Biol. 2011;21:1326–1330. doi: 10.1016/j.cub.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek JH, Cosman P, Feng Z, Silver J, Schafer WR. J Neurosci Meth. 2002;118:9–21. doi: 10.1016/s0165-0270(02)00117-6. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe PE, Chau J, Flanagan KA, Reedy AR, Schriefer LA. Genetics. 2010;184:79–90. doi: 10.1534/genetics.109.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benian GM, Tinley TL, Tang X, Borodovsky M. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. J Mol Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Ferrara TM, Flaherty DB, Benian GM. J Musc Res Cell Motil. 2005;26:435–447. doi: 10.1007/s10974-005-9027-4. [DOI] [PubMed] [Google Scholar]
- 29.Benian GM, Ayme-Southgate A, Tinley TL. Rev Physio, Biochem Pharm. 1999;138:235–268. doi: 10.1007/BFb0119629. [DOI] [PubMed] [Google Scholar]
- 30.Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller RK, Qadota H, Stark TJ, Mercer KB, Wortham TS, Anyanful A, Benian GM. Mol Biol Cell. 2009;20:3608–3616. doi: 10.1091/mbc.E09-03-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. Mol Biol Cell. 2008;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barstead RJ, Kleiman L, Waterston RH. Cell Motil Cytoskel. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- 34.Lecroisey C, Martin E, Mariol MC, Granger L, Schwab Y, Labouesse M, Segalat L, Gieseler K. Mol Biol Cell. 2008;19:785–796. doi: 10.1091/mbc.E07-05-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Jones TI, Tang VW, Brieher WM, Jones PL. J Cell Sci. 2010;123:1116–1123. doi: 10.1242/jcs.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKeown CR, Han HF, Beckerle MC. Devel Dyn. 2006;235:530–538. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han HF, Beckerle MC. Mol Biol Cell. 2009;20:2361–2370. doi: 10.1091/mbc.E08-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaller MD, Borgman CA, Cobb BS, Vines RR, Reynolds AB, Parsons JT. Proc Natl Acad Sci USA. 1992;89:5192–5196. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polet D, Lambrechts A, Ono K, Mah A, Peelman F, Vandekerckhove J, Baillie DL, Ampe C, Ono S. Cell Motil Cytoskel. 2006;63:14–28. doi: 10.1002/cm.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogalski TM, Gilbert MM, Devenport D, Norman KR, Moerman DG. Genetics. 2003;163:905–915. doi: 10.1093/genetics/163.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- 43.Feng Z, Cronin CJ, Wittig JH, Sternberg PW, Schafer WR. BMC Bioinform. 2004;5:515. doi: 10.1186/1471-2105-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng W, Cosman P, Berry CC, Feng Z, Schafer WR. IEEE Trans Biomed Eng. 2004;51:1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- 45.Cronin CJ, Mendel JE, Mukhtaqr S, Kim YM, Stirbl RC, Bruck J, Sternberg PW. BMC Genet. 2005;6:5. doi: 10.1186/1471-2156-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray JM, Hill JJ, Bargmann CI. Proc Natl Acad Sci USA. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. PLoS Comput Biol. 2008;4(4):e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramot D, Johnson BE, Berry TL, Carnell L, Goodman MB. PLoS One. 2008;3(5):e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swierczek NA, Giles AC, Rankin CH, Kerr RA. Nature Meth. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buckingham SD, Sattelle DB. BMC Neurosc. 2009;10:84. doi: 10.1186/1471-2202-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]