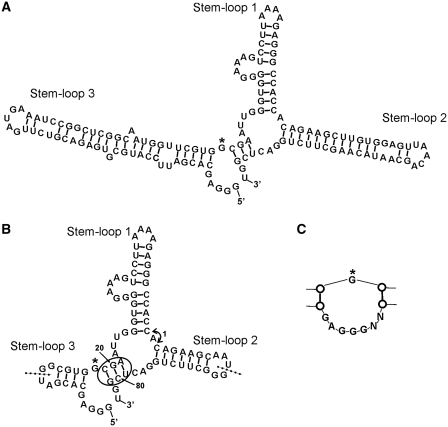
Figure 5.
Confirmed secondary structure of AP3. (A) Schematic diagram of the secondary structure of AP3 (143 nt). Stem–loop 1 was formed by the core sequence from G16. (B) Secondary structure of minimized AP3 (83 nt). Stem–loop 2 and Stem–loop 3 were shortened by truncating from the apex loops of both Stem–loops inward. New random loops were added to the apexes of both minimized stems. The circle indicates site where addition of paired nucleotides between positions 20/21 and 79/80 does not alter binding to GFP, whereas addition of single nucleotide at either site resulted in loss of binding, confirming the stem structure in this region. Insertion of nucleotides between Stem–loop 1 and Stem–loop 2 (arrow 1) also reduced binding to GFP. Dotted lines show other 5′-/3′-ends of fully functional circular permutated aptamers. (C) Re-selection of Stem–loop 3 resulted in reduction of the internal loop (G marked by asterisk in A and B for orientation). Removal of nucleotides within this loop, even a single nucleotide deletion from either the 5′- or 3′-end in the form shown in (A), resulted in loss of function.