Abstract
DNA repeats constitute potential sites for the nucleation of secondary structures such as hairpins and cruciforms. Studies performed mostly in bacteria and yeast showed that these noncanonical DNA structures are breakage-prone, making them candidate targets for cellular DNA repair pathways. Possible culprits for fragility at repetitive DNA sequences include replication and transcription as well as the action of structure–specific nucleases. Despite their patent biological relevance, the parameters governing DNA repeat-associated chromosomal transactions remain ill-defined. Here, we established an episomal recombination system based on donor and acceptor complementary DNA templates to investigate the role of direct and inverted DNA repeats in homologous recombination (HR) in mammalian cells. This system allowed us also to ascertain in a stringent manner the impact of repetitive sequence replication on homology-directed gene repair. We found that nonspaced DNA repeats can, per se, engage the HR pathway of the cell and that this process is primarily dependent on their spacing and relative arrangement (i.e. parallel or antiparallel) rather than on their sequence. Indeed, our data demonstrate that contrary to direct and spaced inverted repeats, nonspaced inverted repeats are intrinsically recombinogenic motifs in mammalian cells lending experimental support to their role in genome dynamics in higher eukaryotes.
INTRODUCTION
The genomes of prokaryotes and eukaryotes harbor numerous and diverse types of repetitive DNA sequences many of which have been associated with genome evolution, regulation of gene expression and chromosomal rearrangements underlying a number of inherited disorders and certain translocation-bearing tumors. These motifs include single direct and inverted DNA repeats with or without internal spacers as well as high-copy-number tandem tracts (1–4). Accumulating evidence indicates that DNA repeats can adopt different noncanonical (i.e. non-B) DNA conformations depending on a number of intrinsic parameters. These include the nucleotide composition, the length and the relative orientation of the constituent DNA units as well as their spacing and extent of sequence identity. Extrinsic factors such as the torsional strain associated with DNA metabolic processes, chromatinization and transcription are also thought to influence the likelihood that DNA repeats acquire higher-order conformations. DNA conformers have been implicated in both physiological and pathological processes including the regulation of DNA replication and expression, oncogenic chromosomal rearrangements and gene amplification (1–7). Related to this, palindromes (i.e. uninterrupted or nonspaced inverted DNA repeats) and inverted DNA repeats with relatively short central spacers, can, via local negative superhelical stress and ensuing intrastrand hybridization and branch migration, extrude into four-way Holliday junction-like DNA structures or cruciforms. Inverted DNA repeats in single-stranded form may also originate stem-loops or hairpins via intrastrand annealing. This may, for instance, occur when the unwinding of double-helical DNA during replication creates a lagging strand template.
The rearrangement of chromosomal DNA carrying noncanonical structures are likely preceded by and dependent on phosphodiester bond cleavage presumably via their resolution and processing by structure-specific nucleases. This might occur in concert with DNA replication-associated phenomena such as replisome stalling or slippage. In Escherichia coli, physical evidence was recently obtained for the emergence of double-stranded DNA breaks (DSBs) at a 246-base pair (bp) palindrome via the combined effects of DNA replication and cleavage by the Mre11/Rad50 homolog SbcCD (8). Interestingly, studies carried out in a yeast model system revealed that inverted repeats of a 320-bp retrotransposon-derived human Alu sequence inserted into a LYS2 reporter allele were a target for the Mre11/Rad50/Nbs1 complex suggesting evolutionary conservation of DNA structure-processing biochemical pathways (9). Moreover, in these prokaryotic and lower eukaryotic model systems, reporter gene expression rescue assays showed that long (i.e. >150 bp) inverted repeat-associated DNA breaks could engage the error-free homologous recombination (HR) pathway (3). Notwithstanding steady progress in this field, many questions remain with respect to the relationships between specific parameters of repetitive DNA motifs, putative ensuing higher-order DNA conformations and the recruitment of cellular pathways that regulate genetic recombination. This knowledge gap is particularly acute in cells of higher eukaryotes (2,3). In addition, hitherto, the vast majority of studies on the biological activity and fate of repetitive DNA in vivo focused on endogenous or exogenous test sequences embedded within the chromosomal DNA of dividing cells. With these types of experimental setups, it is difficult to assess a possible contribution of template DNA replication to repeat-associated phenomena.
Here, we developed and deployed an extrachromosomal recombination system to specifically address the role of single DNA repeats of different sequence, arrangement (i.e. parallel or antiparallel) and spacing in HR-mediated DNA repair in mammalian cells. Furthermore, the introduction of a eukaryotic origin of replication into the repetitive DNA-containing episomes allowed us to also investigate the impact of target template DNA replication on the recombinogenic potential of the various motifs. We demonstrate that simple palindromes and composite inverted DNA repeats, but not direct or spaced inverted DNA repeats, serve as targets for the error-free HR repair pathway in mammalian cells and that this process is independent of ongoing DNA repeat-bearing molecule replication.
MATERIALS AND METHODS
Cells
HeLa cells [American Type Culture Collection (ATCC)], human embryonic kidney (HEK) 293T cells (ATCC) and 911 cells (10) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS; Invitrogen). PER.tTA.Cre76 cells (11) and COS-7 cells (ATCC) were propagated in DMEM supplemented with 10% FBS. All cells were cultured at 37°C in an atmosphere of 10% CO2 in humidified air.
Recombinant DNA
Plasmid pA1.GFP.A2 has been described previously (GenBank accession number: GQ380658) (12). An XmaJI recognition sequence was introduced in pA1.GFP.A2 at nucleotide positions 620 through 625 of the humanized Renilla reniformis green fluorescent protein (GFP) open reading frame (ORF) by polymerase chain reaction (PCR) site-directed mutagenesis to generate the acceptor plasmid pR6K.GFP.STOP. Moreover, pR6K.GFP.STOP has the GFP ORF disrupted by an amber stop codon (Figure 1). The nucleotide sequences of the sense and antisense primers used for introducing the mutation that created the XmaJI site were 5′-GAAGACCTAGGTGGAGGAC-3′ and 5′-GTCCTCCACCTAGGTCTTC-3′, respectively (point mutation is underlined) and the PCR was carried out with Phusion High-Fidelity DNA polymerase (Finnzymes) according to the instructions provided by the manufacturer. Oligodeoxyribonucleotides used for the introduction of DNA sequences into the XmaJI site of pR6K.GFP.STOP were 5′-CTAGGAGCGAGCGAGCGAGCGAGCGAGCGCCGAGCCCCAACTAGT-3′ and 5′-CTAGACTAGTTGGGGCTCGGCGCTCGCTCGCTCGCTCGCTCGCTC-3′ (DR/IR.1), 5′-CTAGGAAGGCGCGAGGGACCGCCGAGCAGGCGAGCCCCAACTAGT-3′ and 5′-CTAGACTAGTTGGGGCTCGCCTGCTCGGCGGTCCCTCGCGCCTTC-3′ (DR/IR.2), 5′-CTAGAGACGACGCAGCGAGCGAGCGAGCGCCACCGACGCACTAGT-3′ and 5′-CTAGACTAGTGCGTCGGTGGCGCTCGCTCGCTCGCTGCGTCGTCT-3′ (DR/IR.3) and 5′-CTAGGAAGGCGCGAGGGAGGGACCGCCGAGCAGGCACCGACGCACTAGT-3′ and 5′-CTAGACTAGTGCGTCGGTGCCTGCTCGGCGGTCCCTCGCGCCTTC-3′ (DR/IR.4). Insertion of a recognition sequence for the meganuclease I-SceI into the XmaJI site of pR6K.GFP.STOP was accomplished using the oligodeoxyribonucleotides 5′-CTAGGAAGTTACGCTAGGGATAACAGGGTAATATAGACTAGT-3′ and 5′-CTAGACTAGTCTATATTACCCTGTTATCCCTAGCGTAACTTC-3′. To generate pR6K.GFP.STOP derivatives containing DNA repeats in a head-to-tail (direct repeat) or tail-to-tail (inverted repeat) configuration, the constructs carrying single copies of the oligodeoxyribonucleotide pairs corresponding to DR/IR.1, DR/IR.2, DR/IR.3 and DR/IR.4 were linearized with BcuI and subsequently subjected to a second round of oligodeoxyribonucleotide cloning. Restriction fragment size analysis was used to distinguish between recombinant plasmids carrying a direct or an inverted repeat of each oligodeoxyribonucleotide pair. To disrupt the palindrome in the IR.1-containing pR6K.GFP.STOP derivative acceptorIR.1 (Figure 1B and C) at the center of symmetry, the plasmid was digested with BcuI and its backbone was combined with the oligodeoxyribonucleotide pair containing the I-SceI recognition sequence (ScR). The resulting construct was designated acceptorspIR.1. To create an acceptor plasmid, in which the GFP ORF is interrupted by the composite adeno-associated virus type 2 (AAV) inverted terminal repeat (ITR), AAV vector shuttle plasmid pDD2 (13) was digested with PvuII and BspLI. The resulting 127-bp AAV ITR-specific DNA fragment was inserted into the XmaJI site of pR6K.GFP.STOP following its blunt-ending with the Klenow fragment of E. coli DNA polymerase I (Klenow, Fermentas) to produce acceptorITR. Plasmid pUC.hrGFPI.SV40pA was prepared by introducing the GFP ORF and the downstream bidirectional simian virus 40 (SV40) polyadenylation signal (pA) derived from pA1.GFP.A2 into pUC19 using BamHI and XbaI. Next, the pA of the rabbit β-globin (βG) gene was inserted immediately upstream of the GFP-coding sequence in pUC.hrGFPI.SV40pA to inhibit possible spurious transcription of the GFP ORF due to the presence of cryptic promoters in the plasmid backbone. To this end, pAAV.hEF1a.DsRedT4.rbGpA, an AAV vector shuttle construct containing a DsRed.T4 (14) expression unit controlled by the human eukaryotic translation elongation factor 1 alpha 1 (EF1α) gene promoter and βG pA, was incubated with NotI and SmiI, the digestion products were blunt-ended using Klenow and the 587-bp βG pA-containing DNA fragment was purified from agarose gel. Subsequently, this fragment was inserted in the proper orientation into pUC.hrGFPI.SV40pA between the Klenow-blunted XbaI and HindIII sites to produce pUC.donor.rbGpA.hrGFPI.SV40pA. Deletion of the GFP start codon from pUC.donor.rbGpA.hrGFPI.SV40pA was achieved by digesting the plasmid with SalI and SdaI, filling in the 3′ recessed ends with Klenow and self-ligation of the plasmid backbone to create the donor template pUC.donor.GFP.ΔATG (i.e. GFPΔATG; GenBank accession number: JF714898). Construct pUC.donor.rbGpA.DsRed.T4.SV40pA was made by replacing the GFP ORF in pUC.donor.rbGpA.hrGFPI.SV40pA by the coding sequence of the red fluorescent protein (RFP) DsRed.T4. The DsRed.T4-coding sequence was excised from pAAV.hEF1a.DsRedT4.rbGpA using XbaI and NotI and subsequently combined with the 3.5-kb XbaI×NotI fragment of pUC.donor.rbGpA.hrGFPI.SV40pA. Disruption of the RFP ORF in pUC.donor.rbGpA.DsRed.T4.SV40pA was accomplished by linearization with NcoI followed by Klenow treatment and self-ligation resulting in nonhomologous donor plasmid pUC.donor.RFP.ΔATG (i.e. RFPΔATG; GenBank accession number: JF714899). The I-SceI expression construct pCAG.I-SceI (GenBank accession number: JF714900) was made by inserting the 3.1-kb SalI×PstI fragment of pCbASce (15) into the polylinker of pUC19 after its digestion with SalI and PstI. The expression plasmid pCAG.I-SceI(Δ112-246) encoding a nonfunctional version of I-SceI was generated by digesting pCAG.I-SceI with BstBI followed by self-ligation of the resulting vector backbone. All DNA preparations were made by using the JetStar 2.0 DNA isolation system (Genomed).
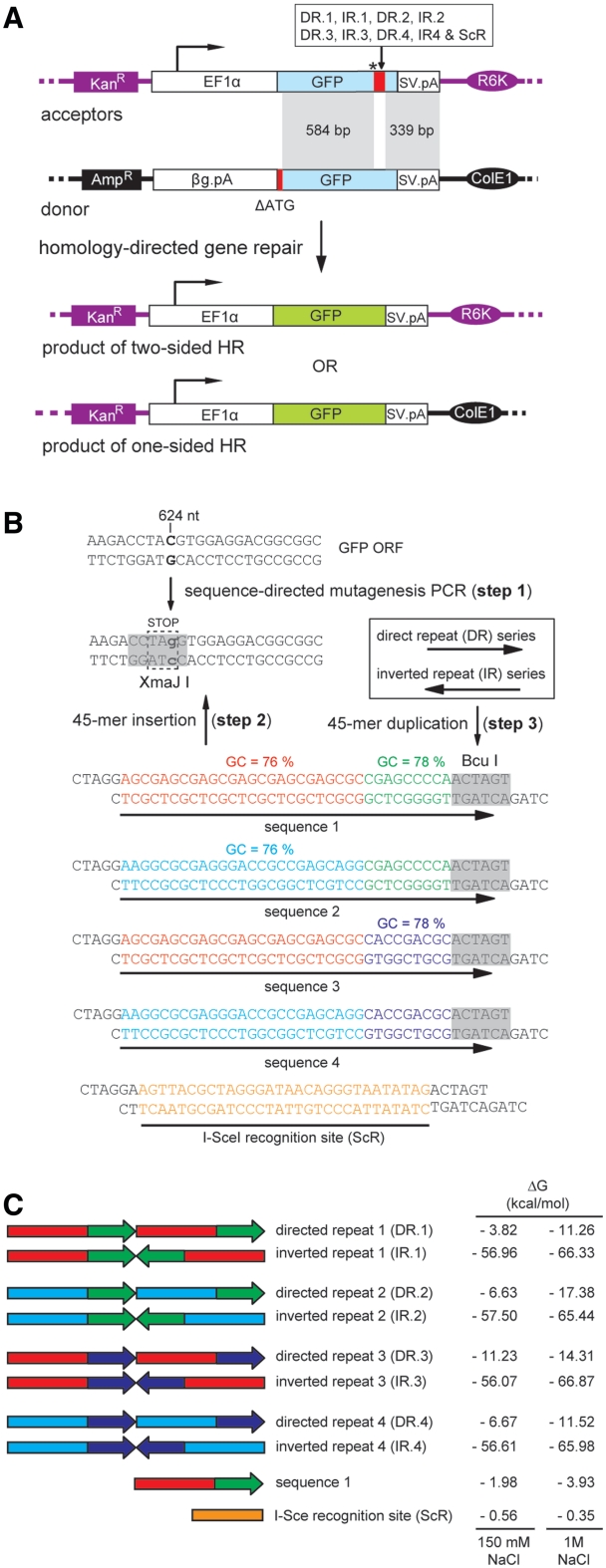
Figure 1.
Episomal recombination system to study the effects of repetitive DNA sequences on homology-directed gene repair in mammalian cells. (A) The bipartite system consists of acceptor and donor plasmids with differently disrupted GFP ORFs. In the acceptor plasmids, the GFP ORF is interrupted by an amber stop codon (asterisk) plus a test DNA sequence of choice (large red bar); whereas in the donor plasmid, it is rendered nonfunctional by the deletion of its first 38 nts (short red bar). The interrupted GFP ORF in the various acceptor plasmids is framed by a constitutively active (broken arrow) human eukaryotic translation elongation factor 1 alpha 1 (EF1α) gene promoter and the SV40 pA (SV.pA), whereas that in the donor plasmid is preceded by the rabbit β-globin pA (βG.pA) and followed by the SV40 pA. The prokaryotic origins of replication R6K and ColE1 as well as the antibiotic resistance genes aminoglycoside 3′-phosphotransferase (KanR) and β-lactamase (AmpR) present in the acceptor and donor plasmid backbones, respectively, are also indicated. Once introduced into cells, donor and acceptor plasmids are candidate substrates for HR by virtue of the shared 339- and 584-bp DNA sequences. Reciprocal exchange of genetic information between acceptor and donor templates via cross-overs within these homologous regions is expected to give rise to transcription units with restored ORFs directing the synthesis of full-length GFP. (B) Three-step strategy to generate the various acceptor plasmids with test sequences 1 through 4 arranged in a direct or inverted repeat orientation (DR and IR series, respectively). Step 1: PCR-based site-directed mutagenesis of the C residue at position 624 of the GFP ORF into a G generates a premature stop codon and a XmaJI recognition site. Step 2: Insertion, at the XmaJI site, of test sequences 1, 2, 3 or 4, which all contain a BcuI recognition sequence at their 3′-end. Step 3: Molecular clones with the BcuI sites in a position distal to the premature stop codon (orientation depicted) were used to duplicate test sequences 1, 2, 3 or 4. Acceptor plasmids with the meganuclease I-SceI recognition site or containing a single copy of target sequence 1 served as controls. (C) Schematic representation of directed and inverted repeats of DNA sequences 1 through 4, the I-SceI recognition site and a single copy of test sequence 1. The propensity of each DNA sequence to transit from lineform to cruciform by intrastrand Watson and Crick base pairing was estimated by calculating the Gibbs free energy (ΔG) in the presence of 150 mM NaCl or of 1M NaCl.
To provide acceptorScR, acceptorDR.1, acceptorIR.1 and acceptorspIR.1 with an SV40 origin of replication (ori), they were linearized with NdeI and blunt-ended using Klenow. Next, these linear DNA molecules were ligated to the SV40 ori-containing 323-bp PvuII×Eco147I fragment of pGL4.22 (GenBank accession number: DQ188842), yielding acceptorScR.ORI, acceptorDR.1.ORI, acceptorIR.1.ORI and acceptorspIR.1.ORI. All oligodeoxyribonucleotides were supplied by Eurofins MWG Operon, while the restriction and DNA modifying enzymes were from Fermentas.
Gibbs free energy calculations
The Gibbs free entry of the most stable secondary structure that can be folded by each of the DNA segments inserted into the XmaJI site of pR6K.GFP.STOP was calculated with the aid of the software program Mfold 3.2 (16) using energy rules for DNA (17) at http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form.
Extrachromosomal DNA extraction
Extrachromosomal DNA was extracted from the transfected cells essentially as described before (18). Briefly, at 72 hours post-transfection, cells were scraped from the surface of a 2-cm2 well with the plunger of a 1-ml Luer-Lok disposable syringe (BD Biosciences). The cell suspension was collected in a 15-ml screwcap tube with a conical bottom (Greiner Bio-One) and centrifuged for 5 min at 1500g. The supernatant was aspirated and the cells were washed once with 5 ml phosphate-buffered saline. After another round of centrifugation, the cell pellet was resuspended in 180 µl of solution I [10 mM Tris–HCl at pH 8.0; 10 mM EDTA; 100 µg/ml proteinase K (Fermentas)] and transferred to a 1.5-ml microtube (Eppendorf). Next, 180 µl of solution II (10 mM Tris–HCl at pH 8.0; 10 mM EDTA; 1.2% sodium dodecyl sulfate) was added. The microtube was inverted thrice to mix its content and incubated for 30 min at 37°C. Next, the sample was mixed with 90 µl of 5 M NaCl and stored overnight at 4°C. The following day, the chromosomal DNA was pelleted by centrifugation for 60 min at 16 100g and the supernatant was removed to a new microtube. Subsequently, the supernatant was extracted twice with buffer-saturated phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform and the episomal DNA was precipitated by the addition of 2.5 volumes of ethanol and 0.5 volumes of 7.5 M ammonium acetate at pH 5.5. After washing with 70% ethanol, the DNA pellet was dried and dissolved in 100 µl of TE+ buffer [10 mM Tris–HCl at pH 8.0; 1 mM EDTA; 100 µg/ml RNase A (Fermentas)]. The purified DNA was used for PCR and Southern blot analyses.
In vivo assay to detect processing of inverted DNA repeats
A total of 80 000 HeLa cells were transfected essentially as described under “DNA transfections” (Supplementary ‘Materials and Methods’ section) with 266 ng of acceptorDR.1, acceptorIR.1 or acceptorScR each mixed with 133 ng of pCAG.I-SceI or of pCAG.I-SceI(Δ112-246). Extrachromosomal DNA was isolated 72 hours post-transfection as described elsewhere in this section and PCR was performed on 4 µl of DNA using 0.4 μM of primers 1 (5′-ATGGTGAGCAAGCAGATCCTGAAG-′3) and 2 (5′-CCGAGAAGGAAGTGCTCC-3′), 0.4 mM of each dNTP (New England Biolabs), 1× GoTaq reaction buffer, 1 mM MgCl2 and 2.5 U of GoTaq DNA polymerase (all from Promega) in a final volume of 50 μl. The PCR cycles were performed in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using the following cycling conditions. A first denaturating step at 95°C for 5 min, followed by 30 cycles of 60 s at 95°C, 60 s at 64°C and 120 s at 72°C. Reactions were terminated by a final extension period of 5 min at 72°C. The synthesized DNA was purified using SureClean (Bioline) and dissolved in 30 µl of 10 mM Tris–Cl, pH 8.0. The resulting PCR products were treated with DpnI alone or with DpnI plus I-SceI, XmaJI or BcuI and, subsequently, subjected to agarose gel electrophoresis. The inclusion of DpnI served to remove possible residual input prokaryotic plasmid DNA before Sourthern blot analysis. The procedures for Southern blotting and for DNA probe radiolabeling are described is the Supplementary ‘Materials and Methods’ section. The probe used is complementary to the GFP ORF and contiguous SV40 pA sequences. Undigested PCR products were cloned in the pCR4-TOPO cloning vector (invitrogen) using GT115 chemically competent E. coli cells. Individual molecular clones corresponding to independent DNA processing events were sequenced using primers 1 or 6 (5′-CAGCTTCGAGGTGGTG-3′).
Statistical analysis
Statistical parameters were computed using Graph Pad Prism 4.03. Student’s t-test was applied to compare data sets with P < 0.05 being considered significant.
RESULTS
Design of the episomal HR assay system
We established an extrachromosomal assay system to study the role of DNA repeats in homology-directed gene repair in vivo. This system is based on pairs of recombination substrates consisting of donor and acceptor (or target) DNA molecules containing GFP reporter genes that are rendered defective by different means (Figure 1A). The transcription unit in the donor plasmid was made nonfunctional by removing the first 38 nucleotides from the GFP ORF and lacks eukaryotic promoter/enhancer elements, whereas the GFP ORF in the various acceptor constructs was disrupted by an internal stop codon preceding the test DNA sequences depicted in Figure 1B and C. The donor and acceptor DNA templates share two regions of perfect sequence identity (Figure 1A, marked in gray), which in the acceptor plasmids are separated from each other by the test sequences. HR-dependent reciprocal exchange of genetic information through the common DNA segments of 584 and 339 bp or via a single cross-over within the upstream, 584-bp, arm of homology are both expected to generate fully functional GFP transcription units. Thus, the capacity of different test DNA sequences to elicit homology-directed gene repair can be readily assessed by the analysis of GFP expression through direct fluorescence microscopy or flow cytometry.
The generation of site-specific DSBs by the Saccharomyces cerevisiae mitochondrial group I intron-encoded homing endonuclease I-SceI at its cognate 18 bp recognition sequence is a well-established method to trigger in a predictable manner DNA repair pathways in prokaryotic and eukaryotic systems (19). Therefore, in our experimental system, we used as positive control for HR-dependent rescue of GFP expression, cells exposed to a donor plasmid, an acceptor template containing the ScR and an I-SceI expression construct. Cells that only received the first two plasmids served as negative control in the HR assay.
Experimental evidence for inverted repeat-dependent formation of DNA secondary structures in acceptor plasmids
The bacteriophage T7 endonuclease I is a commonly used tool to probe for the presence of cruciform- or Holliday-like secondary structures in DNA. This resolvase recognizes preferentially the four-way junction characteristic of these types of DNA conformers and, in its dimeric form, introduces paired nicks close to the branch points leading to subsequent DNA cleavage. On the other hand, negative superhelical torsional stress, such as present in the supercoiled (SC) fraction of plasmid DNA, is a major driving force in the nucleation and extrusion of cruciform-like structures at DNA repeats. Thus, the E. coli enzyme DNA topoisomerase I by catalyzing the relaxation of negatively SC DNA should, to a great extent, inhibit the generation of cruciforms as scored in in vitro assays based on the accumulation of T7 endonuclease I-resolved plasmid DNA molecules (i.e. linear form). The results presented in Figure 2A correspond to an experiment in which the validated cruciform-forming plasmid pUC(AT) was used to confirm that the time-dependent accumulation of the linear T7 endonuclease I-derived DNA product does indeed depend on SC DNA. In fact, in the presence of the DNA topoisomerase I, virtually all the pUC(AT) plasmid transited from the SC to the open circular (OC) form resulting in a concomitant stringent inhibition of cruciform resolution. Importantly, an equivalent experimental outcome was observed when the DNA substrate harboring the test IR.1 sequence was deployed, suggesting that this plasmid is prone to the acquisition of cruciform-like structures as well (Figure 2B).
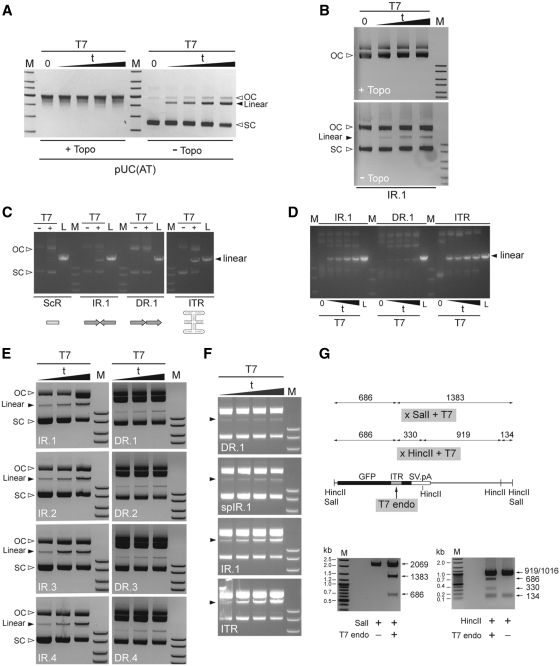
Figure 2.
In vitro assay to probe for the formation of cruciform-like secondary structures in DNA repeat-containing acceptor plasmids. (A) Testing the effect of the negative supercoiling-relaxing enzyme DNA topoisomerase I on the yields of T7 endonuclease I-resolved DNA (solid arrowhead) using the validated cruciform-forming plasmid pUC(AT). pUC(AT) treated or not treated with DNA topoisomerase I (+Topo and −Topo, respectively) was exposed to T7 endonuclease I for 10, 20, 30 or 60 min or underwent a 60-min mock treatment (0). Following agarose gel electrophoresis, the resulting DNA forms were visualized through ethidium bromide staining. OC and SC, open circular and supercoiled DNA forms, respectively. (B) Testing the impact of the negative supercoiling-relaxing enzyme DNA topoisomerase I on the yields of T7 endonuclease I-resolved DNA (solid arrowhead) using the acceptor substrate containing the test nonspaced inverted repeat sequence IR.1. AcceptorIR.1 treated or not treated with DNA topoisomerase I (+Topo and –Topo, respectively) was exposed to T7 endonuclease I for 10, 20 or 30 min or was subjected to a 30-min mock treatment (0). OC and SC, open circular and supercoiled DNA forms, respectively. (C) Target DNA plasmids acceptorScR (ScR), acceptorIR.1 (IR.1), acceptorDR.1 (DR.1) and acceptorITR (ITR) were incubated in the presence (+) or absence (−) of T7 endonuclease I (T7) for 10 min. The resulting DNA products were resolved by agarose gel electrophoresis and stained with ethidium bromide. OC and SC, open circular and supercoiled DNA forms, respectively. L, Acceptor plasmid linearized with ApaLI. (D) Target DNA plasmids acceptorIR.1, acceptorDR.1 and acceptorITR were either incubated with T7 endonuclease I for 10, 20, 30 or 60 min or underwent a 60-min mock treatment (0). The resulting DNA products were analyzed by agarose gel electrophoresis and ethidium bromide staining. L, Acceptor plasmid linearized with ApaLI. (E) The test plasmids harboring the nonspaced inverted repeat sequences IR.1 through IR.4 (left panels) as well as those containing their respective direct repeat counterparts (right panels) were treated with T7 endonuclease I for 20, 30 or 60 min and then resolved through agarose gel electrophoresis. SC, supercoiled DNA; solid arrowheads point to the resolved linear molecules. (F) The plasmids acceptorDR.1, acceptorspIR.1, acceptorIR.1 and acceptorITR were incubated with T7 endonuclease I for 10, 20, 30 and 60 min and analyzed by agarose gel electrophoresis. (G) In vitro mapping of the T7 endonuclease I cleavage site in acceptor molecules. Upper panel, diagram of the expected digestion patterns resulting from the combined activities of SalI and T7 endonuclease I or of HincII and T7 endonuclease I. The numerals correspond to the sizes (in bp) of the different DNA fragments each of which drawn in relation to the parental acceptor DNA template containing the ITR sequence embedded within the GFP ORF. Lower left and right panels, agarose gel electrophoresis of ITR-containing acceptor molecules treated only with SalI or with SalI and T7 endonuclease I or exposed to HincII or to HincII plus T7 endonuclease I, respectively. Lanes M in all the panels, GeneRuler DNA Ladder Mix molecular weight marker (Fermentas).
Next, to investigate the ability of different tandem arrangements of the test DNA sequences to induce the nucleation and extrusion of cruciform-like structures in plasmid substrates, the ScR-, IR.1- or DR.1-containing acceptor constructs were treated or mock-treated with T7 endonuclease I. For the sake of clarity, hereinafter these acceptor plasmids will be named after the test sequences that they contain (e.g. acceptorScR, acceptorIR.1, etc). Because acceptorScR purposely possesses a test DNA sequence with an intrinsically low intrastrand folding capacity (ΔG = −0.35 kcal/mol) it served to establish the background of the assay. To provide for a test DNA sequence known to give rise to four-way DNA structures under physiological conditions, we generated acceptorITR, which has a 127-bp DNA segment derived from the AAV ITR inserted at GFP nucleotide position 624. This DNA segment, which contains three self-complementary regions (i.e. A/A′, B/B′ and C/C′), has a high propensity to fold into a T-shaped hairpin structure by intrastrand hybridization (20,21). Thus, the acceptorITR plasmid was also subjected to T7 endonuclease I treatment. Furthermore, to generate internal references, each acceptor plasmid was linearized in parallel with the restriction enzyme ApaLI.
Agarose gel electrophoresis of T7 endonuclease I- or ApaLI-treated acceptor plasmids revealed that the fragments with sizes consistent with DNA cleavage (i.e. linearized DNA forms) were clearly more prominent in the samples of acceptorIR.1 and acceptorITR than in those corresponding to acceptorScR and acceptorDR.1 (Figure 2C). Indeed, there was no noticeable difference in the accumulation of linear DNA between the non-repeat-containing acceptorScR and the direct repeat-containing acceptorDR.1. As expected, agarose gel electrophoresis of acceptor plasmids not exposed to T7 endonuclease I resulted in the detection of only SC and OC (i.e. nicked) DNA topologies (Figure 2C).
In another set of experiments, we investigated the T7 endonuclease I-dependent accumulation of linear acceptor DNA species as a function of time. To this end, constructs acceptorIR.1, acceptorDR.1 and acceptorITR were incubated with the resolvase for 10, 20, 30 or 60 min or left untreated. Analysis of the resulting DNA products by agarose gel electrophoresis showed that for each period of T7 endonuclease I treatment, the inverted repeat-containing plasmids acceptorIR.1 and acceptorITR yielded higher amounts of linear DNA than the direct repeat-containing construct acceptorDR.1 (Figure 2D). In fact, exposure of acceptorDR.1 for 60 min to T7 endonuclease I generated less linear DNA molecules than a 10-min incubation of acceptorITR or acceptorIR.1 with the same enzyme (Figure 2D). However, linear acceptorITR molecules accumulated with faster kinetics than linear acceptorIR.1 DNA (Figure 2D). These data, in agreement with the calculated folding free energy values (Figure 1C), indicate that T7 endonuclease I-susceptible secondary DNA structures (e.g. DNA cruciforms) can be formed after insertion of the test DNA sequences as inverted repeats in acceptor plasmid substrates (Figure 1C). These experimental results further suggest that the AAV ITR has a higher propensity to acquire a T7 endonuclease I-sensitive DNA conformation than IR.1. Similar experiments were also carried out with not only the acceptor substrates harboring IR.1, DR.1 and ITR but also with those containing each of the other six nonspaced repeat sequences (i.e. DR.2, DR.3, DR.4, IR.2, IR.3 and IR.4) as well as that with the spaced inverted repeat sequence spIR.1. Results from these experiments established that for each of the IR/DR acceptor pairs those substrates containing the nonspaced inverted repeat led, at very time point analyzed, to a higher accumulation of T7 endonuclease I-resolved linear forms than that resulting from those harboring the nonspaced direct repeat (Figure 2E, compare each left panel with the corresponding right panel). Of note, acceptorspIR.1 with its spaced inverted repeat was clearly less prone to T7 endonuclease I digestion than its acceptorIR.1counterpart (Figure 2F, compare second with third panel from the top). Once again, acceptorITR with its multi-palindromic AAV ITR sequence displayed a somewhat higher susceptibility to the T7 endonuclease I when compared with that of acceptorIR.1 (Figure 2F, compare the two lowest panels). Finally, to identify the position corresponding to the major T7 endonuclease I cleavage site in acceptor DNA backbones with secondary structure-forming test sequences, we incubated an ITR-containing construct (Figure 2G, upper panel) exclusively with SalI or with SalI and T7 endonuclease I or exposed it to HincII or HincII and T7 endonuclease I. Agarose gel electrophoresis of the resulting DNA fragments revealed digestion patterns fully consistent with T7 endonuclease I-dependent cleavage of the acceptor templates at the location of the inverted repeat (Figure 2G, lower panels).
Inverted repeats stimulate DNA exchange through HR in mammalian cells
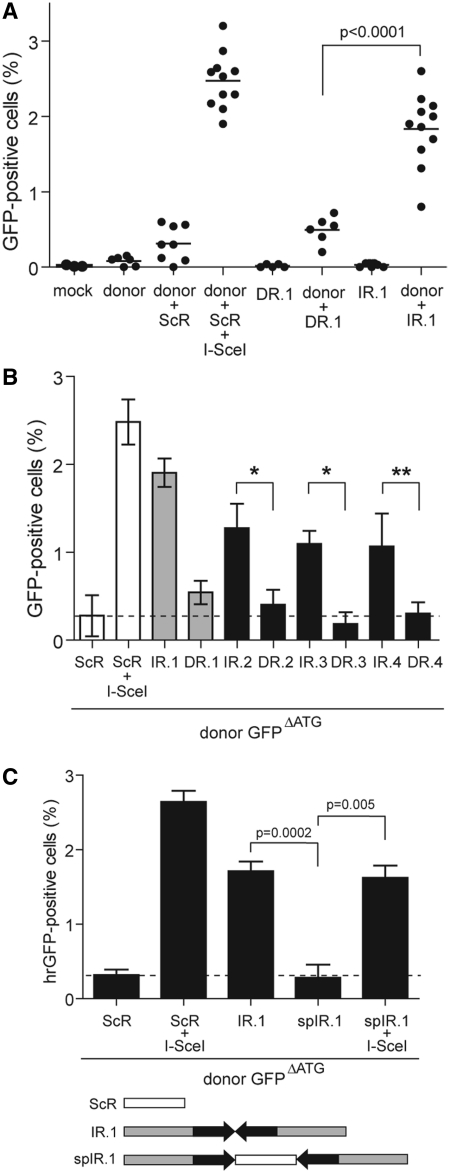
Next, we deployed the aforementioned episomal HR assay system (Figure 1) to ask whether DNA templates containing direct or inverted DNA repeats can serve as substrates for homology-directed gene repair in mammalian cells. In these experiments, HeLa cells were either mock-transfected or transfected with different plasmid combinations. At 4 days post-transfection, HR-dependent GFP repair was measured by flow cytometry. Transfection of the donor construct GFPΔATG, acceptorDR.1 or acceptorIR.1 alone did not give rise to measurable GFP-specific signals showing that the mutations introduced in these plasmids did functionally disrupt the GFP ORF (Figure 3A). Cotransfection of both GFPΔATG and acceptorScR resulted in a low percentage of GFP-positive cells [i.e. 0.3 ± 0.2% (n = 8); Figure 3A, donor + ScR], establishing the background of the assay in the presence of both donor and acceptor templates. To validate the extrachromosomal recombination system, we relied on I-SceI-mediated site-specific DSB formation, which is a well-established method to induce HR in a controlled and predictable manner [see e.g. ref. (19)]. For this purpose, HeLa cells were cotransfected with GFPΔATG, acceptorScR and the I-SceI-encoding expression plasmid pCAG.I-SceI. The inclusion of pCAG.I-SceI resulted in a large increase in the frequency of GFP-positive cells [i.e. 2.5 ± 0.4 % (n = 11); Figure 3A, donor + ScR + I-SceI]. Interestingly, while cotransfection of GFPΔATG and acceptorDR.1 yielded background levels of GFP-positive cells (Figure 3A, compare donor + ScR with donor + DR.1), codelivery of the donor construct together with acceptorIR.1 gave rise to a significantly higher percentage of GFP-positive cells [i.e. 1.9 ± 0.5 % (n = 11); Figure 3A, compare donor + ScR and donor + DR.1 with donor + IR.1]. These data imply that a tandem of test sequence 1, when arranged in an inverted repeat orientation, serves as an effective target for homology-directed gene repair.
Figure 3.
Effect of repetitive DNA sequences in a direct or inverted repeat configuration on homology-directed gene repair in HeLa cells. (A) HeLa cells were transfected with acceptorDR.1 or acceptorIR.1 alone (DR.1 and IR.1, respectively) or with either of these two acceptor plasmids in combination with the donor construct GFPΔATG (donor + DR.1 and donor + IR.1, respectively). Mock-transfected HeLa cells (mock) and HeLa cells transfected with GFPΔATG alone (donor) or together with acceptorScR (donor + ScR) served as negative controls. The positive control for the rescue of GFP expression by HR was provided by cotransfecting HeLa cells with acceptorScR, GFPΔATG and the I-SceI-encoding plasmid pCAG.I-SceI (donor + ScR + I-SceI). Quantification of the number of GFP-positive cells was carried out by flow cytometry at 4 days post-transfection. A minimum of 5 and a maximum of 11 independent experiments were performed with 10 000 events corresponding to viable cells being measured per sample. (B) HeLa cells were cotransfected with GFPΔATG plus either acceptorIR.2, acceptorDR.2, acceptorIR.3, acceptorDR.3, acceptorIR.4 or acceptorDR.4. To facilitate comparison, data sets corresponding to HeLa cells cotransfected with GFPΔATG and acceptorScR or with GFPΔATG, acceptorScR and pCAG.I-SceI as well as those corresponding to HeLa cells cotransfected with GFPΔATG and either acceptorIR.1 or acceptorDR.1 presented in Figure 3A are repeated in Figure 3B (open and gray bars, respectively). Quantification of GFP expression rescue was carried out by flow cytometry at 4 days post-transfection. Cumulative data from 4 different experiments (solid bars) are expressed as mean ± standard deviation. *P = 0.002, **P = 0.009. (C) Flow cytometric analysis of HeLa cells that, in addition to being exposed to the donor plasmid GFPΔATG also, received acceptorScR, acceptorIR.1 or acceptorspIR.1. In the latter construct, the inverted repeat of test sequence 1 is interrupted at its axis of symmetry by an I-SceI recognition site (see diagram below the graph). HeLa cells cotransfected with GFPΔATG, the I-SceI encoding plasmid pCAG.I-SceI and either acceptorScR or acceptorspIR.1 served as positive controls for HR-mediated GFP repair. Data corresponding to a minimum of three different experiments are shown as mean ± standard deviation.
To further investigate the relationship between the relative orientation of repetitive DNA sequences and HR-mediated gene repair, HeLa cells were cotransfected with GFPΔATG and with an acceptor plasmid containing a tandem of test sequences 2, 3 or 4 arranged in either a direct or in an inverted repeat configuration. Consistent with the previous results (Figure 3A), acceptor plasmids endowed with the various inverted repeats yielded significantly higher numbers of GFP-positive cells than their isogenic direct repeat-containing counterparts, which gave rise to frequencies of GFP-positive cells not significantly above background level (i.e. GFPΔATG + acceptorScR; Figure 3B). Thus, at least for the four different test DNA sequences that were investigated, which possess the same GC content; induction of HR is primarily dependent on the arrangement of the repetitive DNA unit as opposed to their specific nucleotide sequence.
Central spacing abolishes inverted DNA repeat-dependent homology-directed gene repair
To study the impact of repetitive DNA spacing in inverted repeat-induced HR, we generated acceptorspIR.1. This plasmid was made by inserting at the axis of symmetry of IR.1, a 42-bp sequence encompassing the ScR to effectively separate test sequence 1 from its reverse complement copy. In these experiments, HeLa cells were transfected with donor plasmid GFPΔATG, mixed with acceptorScR, acceptorIR.1 or acceptorspIR.1 and, where indicated, pCAG.I-SceI. As previously observed, cotransfection of GFPΔATG and acceptorIR.1 resulted in significant HR-dependent GFP expression (Figure 3C). However, substitution of acceptorIR.1 by acceptorspIR.1 yielded GFP expression rescue activity levels that were not above those detected in cell cultures exposed to GFPΔATG and acceptorScR (Figure 3C). Possibly, physical separation of the repetitive DNA unit as in acceptorspIR.1 inhibits the in vivo formation of cruciform-like structures as suggested by our in vitro assay results (Figure 2F) as well as those of others (22–24), which renders the transfected DNA templates no longer a target for the HR machinery. Addition of pCAG.I-SceI to the transfection mixtures consisting of GFPΔATG and either acceptorScR or acceptorspIR.1 rescued high-level GFP expression (Figure 3C). The latter outcome shows that acceptorspIR.1 does not contain an intrinsically defective GFP target template. Taken together, these data indicate that perfect palindromes or nonspaced inverted DNA repeats are preferred over spaced inverted DNA repeats as targets for homology-directed gene repair in vivo presumably due to their capacity to form secondary structures in vivo that can subsequently serve as direct targets for cellular structure-specific nucleases.
Experimental evidence for in vivo nuclease-mediated processing of nonspaced inverted DNA repeats
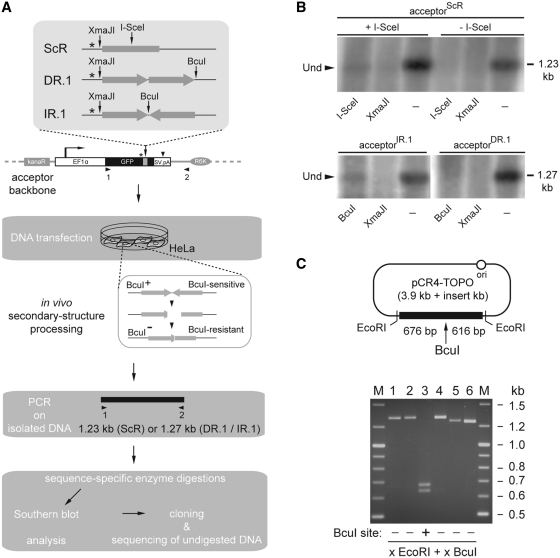
In the search of evidence for nuclease-mediated processing of palindromic test sequences in vivo, we set-up the assay system illustrated in Figure 4A. In this assay, HeLa cells are transfected with acceptorIR.1, acceptorDR.1 or with acceptorScR mixed with pCAG.I-SceI or with pCAG.I-SceI(Δ112-246). Expression plasmid pCAG.I-SceI(Δ112-246) encodes a nonfunctional I-SceI protein. Cells cotransfected with acceptorScR and pCAG.I-SceI constitute a positive control for in vivo site-specific DSB formation at acceptor templates. A key feature of this assay is the fact that a discriminating marker in the form of a BcuI recognition site lies at the axis of symmetry of the test nonspaced inverted repeat sequence (Figures 1B and 4A, upper panel). Generation of cruciforms at this sequence followed by its recognition and processing by cellular structure-specific nuclease(s) should result in DNA breaks. The resulting DNA can subsequently serve as a substrate for error-prone DNA repair processes in the cell, such as nonhomologous end-joining (NHEJ), eventually leading to the emergence of a population of BcuI-resistant acceptor molecules (Figure 4A). Similarly, processing of I-SceI-mediated DSBs by a cellular error-prone DNA repair pathway should yield templates that are knocked-out in the I-SceI cognate target site. Thus, DNA processing/repair at specific test sequences should lead to a mixture of GFP templates that can be PCR-amplified and discriminated on the basis of sequence-specific enzymatic digestions combined with Southern blot and nucleotide sequence analysis. Southern blotting of amplicons made with the aid of primer set 1/2 (Figure 4A), showed the presence of I-SceI-undigested templates in extrachromosomal DNA isolated from HeLa cells cotransfected with acceptorScR and pCAG.I-SceI (+I-SceI; Figure 4B) but not in those cotransfected with acceptorScR and pCAG.I-SceI(Δ112-246) (-I-SceI; Figure 4B). Importantly, the same analysis applied to episomal DNA extracted from HeLa cells transfected with acceptorIR.1 or with acceptorDR.1 revealed the presence of BcuI-undigested templates in cells exposed to the former construct (Figure 4B, lower-left panel). The fact that PCR products amplified from acceptorScR or from acceptorIR.1 and treated with XmaJI did not yield discernable undigested material suggests that the majority of the DNA sequence modifications took place in the vicinity of the respective I-SceI site or of the IR.1 axis of symmetry. Next, amplicons isolated from the BcuI-undigested fraction were cloned into pCR4-TOPO and subjected to restriction fragment length analysis using BcuI. This analysis confirmed the disruption of the BcuI restriction site in several of the analyzed clones (Figure 4C, lanes 1, 2, 4, 5 and 6). Moreover, the apparently different molecular weights of the restriction fragments migrating slower than 1.2 kb-sized linear DNA suggest that the BcuI-refractory clones do harbor sequences representing the end-product of independent DNA processing/repair events. To confirm this and to identify the breakpoints in acceptorIR.1 templates at the nucleotide level, we carried out DNA sequence analysis of clones 5 and 6 (Supplementary Figure S1). As a control, we also sequenced a pCR4-TOPO-based clone corresponding to PCR-amplified DNA from HeLa cells cotransfected with acceptorScR and pCAG.I-SceI (Supplementary Figure S1).
Figure 4.
In vivo processing and repair of nonspaced inverted DNA repeats. (A) Diagrammatic representation of the experimental set-up deployed to study the processing and repair of nonspaced inverted DNA repeats in cells (see text for details and legend of Figure 1A for an explanation of the symbols). (B) Southern blot analysis using an GFP ORF- and SV40 pA-specific probe of PCR products derived from extrachromosomal DNA isolated from HeLa cells cotransfected with acceptorScR and pCAG.I-SceI (+I-SceI), acceptorScR and pCAG.I-SceI(Δ112-246) (-I-SceI), acceptorIR.1 or acceptorDR.1. Before electrophoresis, the DNA samples were treated with DpnI alone (−) or with DpnI together with I-SceI, XmaJI or BcuI. Sizes corresponding to undigested amplicons (solid arrowhead) expected for the primer pair 1/2 are indicated on the right of the autoradiograms. (C) Upper panel, Diagram of a pCR4-TOPO molecular clone containing a PCR product amplified from extrachromosomal DNA isolated from HeLa cells transfected with acceptorIR.1 and treated with BcuI. Vertical arrow points to the original position of the BcuI recognition site. Lower panel, Agarose gel electrophoresis of BcuI-treated pCR4-TOPO clones harboring PCR products amplified from episomal DNA extracted from HeLa cells transfected with acceptorIR.1 and exposed to BcuI digestion (lanes 1 through 6). Lane M, GeneRuler DNA Ladder Mix molecular weight marker.
Inverted DNA repeat-dependent homology-directed gene repair occurs in a variety of mammalian cell types
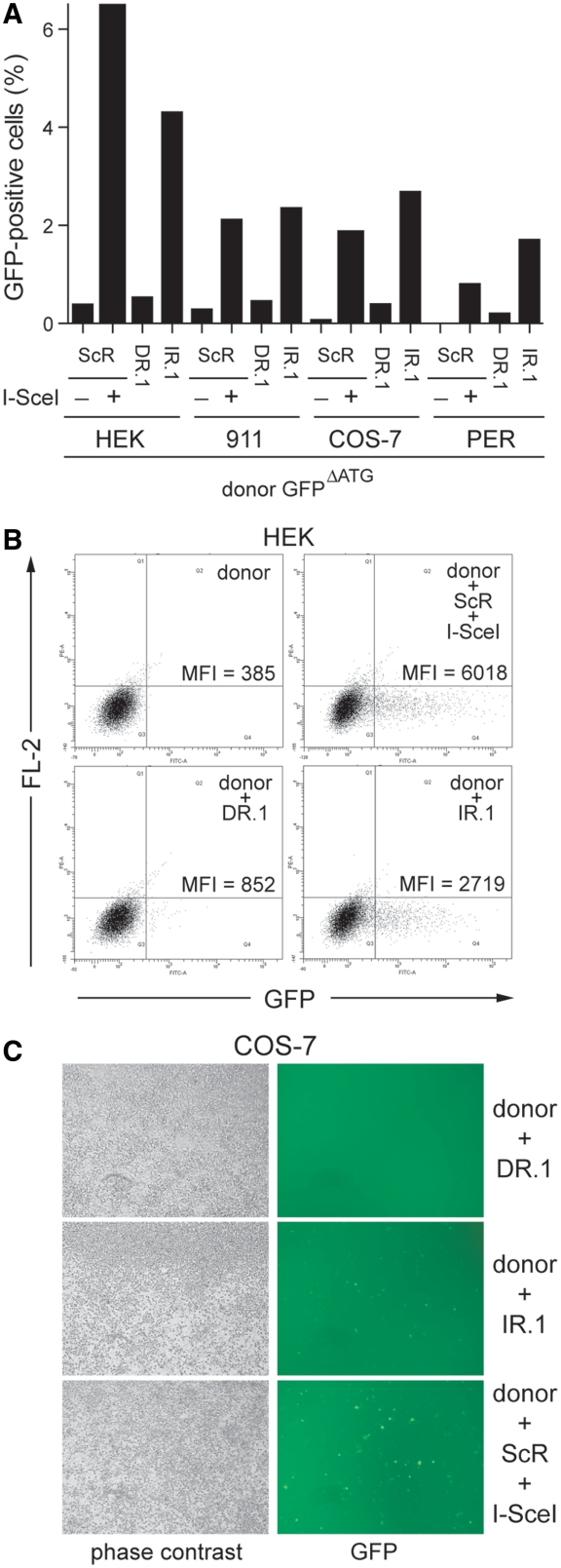
Subsequently, we exposed cultures of HEK 293T cells, human fetal retinoblasts (911 and PER.tTA.Cre76 cells) and African green monkey kidney fibroblasts (COS-7 cells) to GFPΔATG and either acceptorDR.1 or acceptorIR.1. Again, negative and positive controls were provided by cotransfecting cultures of each of these cell types with a mixture of the donor plasmid GFPΔATG and acceptorScR alone or together with pCAG.I-SceI, respectively. Data depicted in Figure 5A show distinct levels of HR-dependent GFP repair in the various cell types tested. This might be the result of different transfection efficiencies and/or of intrinsic cell type-specific differences in the ability to recognize and process, via HR, DNA secondary structures. However, importantly, like in HeLa cells, appreciable HR-mediated GFP repair was only observed after cotransfection of GFPΔATG and acceptorIR.1 (Figure 5A). This IR.1-mediated HR stimulatory effect was independent of the amount of p53 present in the disparate cell types tested (Supplementary Figure S2). Some representative flow cytometry dot plots and direct fluorescence microscopy micrographs corresponding to these experiments are depicted in Figure 5B and C, respectively. Collectively, these experiments suggest that the inverted DNA repeat-induced HR is a mammalian cell type-independent phenomenon.
Figure 5.
Effect of repetitive DNA sequences in a direct or inverted repeat configuration on homology-directed gene repair in mammalian cells. (A) Flow cytometric analysis of human embryonic kidney 293T cells (HEK), human fetal retinoblasts (911 and PER.tTA.Cre76 [PER]) and African green monkey kidney fibroblasts (COS-7) cotransfected with GFPΔATG and either acceptorDR.1 or acceptorIR.1. Negative and positive controls for HR-mediated GFP repair in each of the tested cell types were provided by cells containing GFPΔATG and acceptorScR (−) or these two plasmids as well as pCAG.I-SceI (+), respectively. (B) Dot plot representation of GFP expression in human embryonic kidney 293T cells (HEK) transfected with GFPΔATG alone (donor) or with a mixture of GFPΔATG and either acceptorDR.1 or acceptorIR.1. Cultures cotransfected with GFPΔATG, acceptorScR and the I-SceI expression plasmid pCAG.I-SceI served as positive control. Flow cytometry was carried out 3 days post-transfection with 10 000 viable cells being analyzed per sample. (C) Live-cell imaging by phase-contrast and fluorescence microscopy of monolayers of African green monkey kidney fibroblasts (COS-7) cotransfected with GFPΔATG and either acceptorIR.1 or acceptorDR.1. Parallel cultures exposed to GFPΔATG, acceptorScR and pCAG.I-SceI served as positive control for HR-dependent GFP reconstitution. Microscopic analysis was performed 3 days post-transfection. Original magnification: ×40.
Composite inverted DNA repeats are equally effective at stimulating homology-directed gene repair as simple palindromes
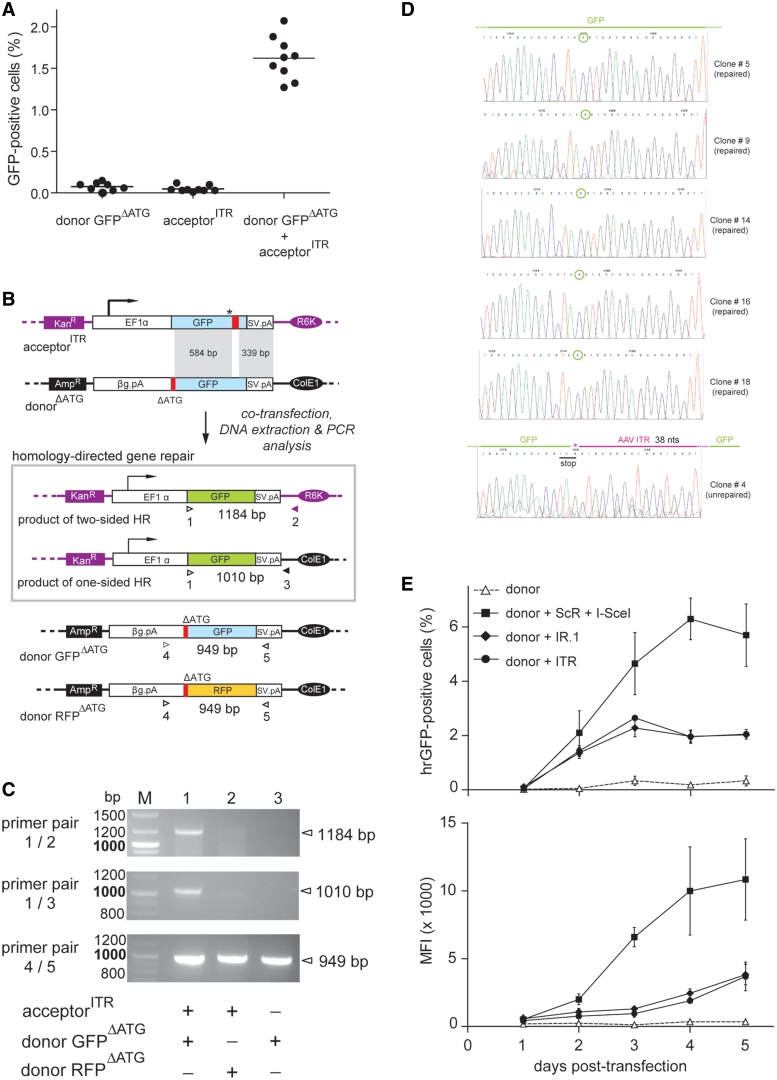
To investigate the capacity of a structurally more ‘complex’ or composite inverted DNA repeat to stimulate HR, we deployed acceptorITR. Contrary to HeLa cell cultures that were transfected with donor GFPΔATG or acceptorITR alone, those exposed simultaneously to GFPΔATG and acceptorITR readily revealed the presence of GFP-positive cells (Figure 6A). The percentages of these cells were similar to those observed in HeLa cell cultures upon cotransfection of GFPΔATG and acceptorIR.1 (Figure 3). To rule out the possibility that DNA sequences prone to secondary structure formation, such as the AAV ITR, alter plasmid DNA transfection efficiency, we transfected HeLa cells with pDsRed or with pDsRed2X.ITR. The former construct contains an expression unit based on the human EF1α promoter, the DsRed.T4 reporter and the rabbit β-globin pA, whereas the latter has this transcription unit flanked by AAV ITRs. Flow cytometric analysis at 72 hours post-transfection showed that cultures exposed to pDsRed2X.ITR had a frequency of reporter-positive cells as well as an amount of DsRed.T4 protein very similar to those measured in cultures transfected with pDsRed. We conclude that the ITR sequences do not significantly affect the transfection efficiency of plasmid DNA (Supplementary Figure S3).
Figure 6.
Comparison of the ability of simple and composite inverted DNA repeats to trigger homology-directed gene repair in mammalian cells. (A) HeLa cells were transfected with GFPΔATG or with acceptorITR or were cotransfected with GFPΔATG plus acceptorITR. Analysis of GFP expression by flow cytometry was carried out 4 days post-transfection on 10 000 viable cells per sample. Horizontal lines representing means corresponding to nine independent experiments. (B) Schematic representation of the PCR-based assay deployed to detect GFP ORFs repaired via homology-directed gene targeting. Primers 1 and 2 were designed to amplify templates resulting from two-sided HR whilst oligodeoxyribonucleotides 1 and 3 were used to specifically detect products of one-sided HR. Primer 1 recognizes the first 24 nts of the GFP ORF. Oligodeoxyribonucleotides 2 and 3 target sequences exclusively present in the acceptor and donor plasmid backbones, respectively. The PCR products of primers 4 and 5, which bind to the rabbit β-globin pA (βG.pA) and SV40 pA (SV.pA), respectively, served as internal control for extrachromosomal DNA quality and quantity. Amplicon size (in bp) expected for each primer pair is indicated. For an explanation of the other symbols see the legend of Figure 1A. (C) PCR analysis using primer pairs 1/2, 1/3 and 4/5 (upper, middle and lower panels, respectively) of extrachromosomal DNA isolated from HeLa cells cotransfected with acceptorITR and the homologous donor plasmid GFPΔATG (lane 1) or with acceptorITR and the nonhomologous donor plasmid RFPΔATG (lane 2) or from HeLa cells transfected with GFPΔATG alone (lane 3). Lane M, GeneRuler DNA Ladder Mix molecular weight marker. (D) Nucleotide sequence data of individual clones corresponding to PCR products obtained with primers 1 and 2. Clones #5, #9, #14, #16 and #18 represent products of HR containing a repaired GFP ORF. The encircled ORF-correcting cytosine is derived from the donor plasmid. Clone #4 corresponds to a rearranged acceptor template featuring 38 bp of the originally introduced AAV ITR (purple line above graph) and retaining the engineered stop codon. The G marked with the asterisk is derived from the original acceptor template. (E) Flow cytometric analysis at the indicated time points of HeLa cells transfected with the donor construct GFPΔATG alone or together with either acceptorIR.1, acceptorITR or a mixture of acceptorScR and the I-SceI-encoding plasmid pCAG.I-SceI. Both the frequencies of GFP-positive cells (upper graph) as well as their average mean fluorescence intensities (MFI; lower graph) are presented.
To confirm, at the molecular level, the accurate repair of GFP ORFs following inverted repeat-mediated HR, we performed PCR analysis on extrachromosomal DNA isolated from HeLa cells transfected with GFPΔATG alone or together with acceptorITR. Extrachromosomal DNA extracted from HeLa cells exposed to acceptorITR in combination with the nonhomologous donor construct RFPΔATG served as an extra negative control. The PCR assay shown in Figure 6B is based on primer pairs 1/2 and 1/3 to amplify PCR products that are diagnostic for the generation of GFP templates corrected by two-sided and one-sided HR, respectively. Although primers 1 and 2 also bind to the acceptor plasmid, their use did not yield any PCR products (Figure 6C) most likely due to the inability of the thermostable polymerase to read through the AAV ITR. As shown in Figure 6C (upper and middle panels), PCR fragments corresponding to reconstituted GFP ORFs were exclusively detected in the DNA sample from the cells that were cotransfected with donor GFPΔATG and acceptorITR. Moreover, the results demonstrate that gene repair was brought about by two-sided as well as by one-sided HR (Figure 6C, lane 1 of upper panel and lane 1 of middle panel, respectively). Of note, amplification reactions carried out on an in vitro mixture of GFPΔATG and acceptorITR plasmids did not yield any product, showing that the detection of specific amplicons in the DNA sample derived from HeLa cells cotransfected with GFPΔATG and acceptorITR was not an artifact but the result of genetic information exchange in vivo (not shown). Internal control PCR amplifications using primers 4 and 5 showed the presence of the homologous and the nonhomologous donor templates GFPΔATG and RFPΔATG, respectively, confirming the integrity of the extrachromosomal DNA following the isolation procedure (Figure 6C, lower panel). Next, we performed an independent transfection experiment in HeLa cells followed by the same DNA isolation procedure and PCR assay. However, in this new experiment, an extra control was included. This consisted in using extrachromosomal DNA from cells transfected with acceptorITR mixed, before PCR, with extrachromosomal DNA from cells transfected with donor GFPΔATG. Data depicted in Supplementary Figure S4 shows, once again, the presence of the specific 1.2-kb amplicon exclusively in the sample corresponding to cells cotransfected with donorΔATG and acceptorITR.
The PCR products obtained with the aid of primer pair 1/2 were inserted into a plasmid vector after which, nucleotide sequence analysis of 20 randomly selected DNA clones was carried out. From this analysis, it has resulted that 19 of these clones contained GFP ORFs without any mutations linking them to error-free HR events (Figure 6D, the 5 uppermost nucleotide sequences serve as examples of this analysis). The remaining clone had the GFP ORF disrupted at the initially engineered premature stop codon by AAV ITR-derived, heterologous DNA (Figure 6D; lowest nucleotide sequence) suggesting that it was the product of inverted repeat microhomology-directed recombination (25) or of error-prone NHEJ, possibly following center-break palindrome revision or cruciform resolution (2,3,26,27). Additionally, we studied the kinetics of homology-directed gene repair involving acceptorITR and acceptorIR.1 and directly compared it with that of the conventional DSB-induced HR. To this end, HeLa cells were transfected with donor GFPΔATG alone or together with acceptorIR.1, acceptorITR or a mixture of acceptorScR and pCAG.I-SceI. Results shown in Figure 6E reveal a time-dependent increase in the number of GFP-positive cells (Figure 6E, upper graph) and in the amount of reporter protein per GFP-positive cell (Figure 6E, lower graph) in all cultures cotransfected with acceptor plasmids and the donor GFPΔATG construct. Interestingly, the time-dependent increase in the frequency and fluorescence intensity of GFP-positive cells was faster in cultures exposed to acceptorScR and pCAG.I-SceI than in those incubated with acceptorITR or with acceptorIR.1. Moreover, no significant differences in both of the GFP-specific parameters were found at all time points tested in cell cultures exposed to GFPΔATG and either acceptorITR or acceptorIR.1 (Figure 6E). We postulate that the lower HR-inducing activity of the inverted DNA repeat sequences when compared with DSBs may relate to their transient nature. Perhaps, secondary structures formed in vivo by certain inverted repeats and palindromes constitute ‘facultative’ or ‘intermittent’ DNA lesions leading to a sporadic engagement of the HR machinery. Another contributing factor may be the larger number of biochemical reactions necessary to process cruciform-like structures by HR than that that is necessary to repair DSBs.
Replication of DNA repeat-containing acceptor molecules does not significantly affect homology-directed gene repair
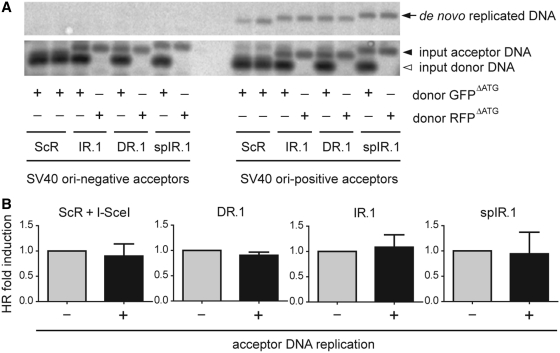
SV40 is a mammalian double-stranded DNA virus with a circular genome whose replication has been extensively studied as a model for chromosomal nuclear DNA replication in higher eukaryotes (28). The only viral cis-acting element and trans-acting factor required for SV40-dependent DNA replication are the ori and the large T antigen protein, respectively. Thus, to investigate the impact of target template replication on DNA repeat-induced HR in mammalian cells, the SV40 ori was introduced at equivalent positions in acceptorScR, acceptorDR.1, acceptorIR.1 and acceptorspIR.1 to generate the constructs acceptorScR.ORI, acceptorDR.1.ORI, acceptorIR.1.ORI and acceptorspIR.1.ORI, respectively. Next, each of these plasmids was individually transfected into the SV40 large T-expressing COS-7 cells together with the homologous donor construct GFPΔATG or the nonhomologous donor plasmid RFPΔATG. Extrachromosomal DNA isolated from these cells was treated with DpnI to selectively digest the input prokaryotic DNA and with XbaI to linearize de novo synthesized acceptor DNA molecules and HR products. Southern blot analysis of the digestion products using a GFP-specific probe revealed SV40 ori-dependent accumulation of de novo generated DNA molecules, demonstrating the replication proficiency of the SV40 ori-containing acceptor plasmids in COS-7 cells (Figure 7A). Interestingly, acceptor DNA replication did not lead to a significant increase in homology-directed gene repair levels in any of the experimental setups tested (Figure 7B). Taken together, these data indicate that under the prevailing experimental conditions, replication of acceptor DNA molecules carrying a palindrome, a direct repeat or a spaced inverted repeat does not significantly enhance homology-directed gene repair.
Figure 7.
Testing the impact of target DNA synthesis on DNA repeat-mediated homology-directed gene repair. (A) SV40 ori-dependent DNA replication of acceptor constructs. Acceptor plasmids containing the test sequences ScR, IR.1, DR.1 or spIR.1 and with or without SV40 ori were transfected into COS-7 cells together with the homologous donor construct GFPΔATG or the nonhomologous donor plasmid RFPΔATG. At 3 days post-transfection, extrachromosomal DNA was extracted and treated with XbaI and the prokaryotic DNA methylation pattern-sensitive restriction enzyme DpnI. After agarose gel electrophoresis, the resolved DNA was subjected to Southern blot analysis using a GFP-specific probe. DpnI-resistant, de novo replicated DNA, was detected only in samples of cells transfected with SV40 ori-positive acceptor plasmids (right-hand side upper panel). (B) Relative homology-directed gene repair frequencies in COS-7 cells transfected with GFPΔATG, the indicated acceptor plasmids with (+) or without (−) SV40 ori and in one case also pCAG.I-SceI.
DISCUSSION
Repetitive DNA sequences include not only single, direct and inverted repeats (2–4), like the ones investigated in this study, but also high-copy-number repetitive DNA tracts such as those corresponding to 1–4 bp microsatellites and 6–64 bp minisatellites (1). Microsatellites include the expandable trinucleotide repeats associated with neurodegenerative and neuromuscular disorders such as Huntington’s and occulopharyngeal muscular dystrophy. Despite their diversity, diverse lines of evidence point to the acquisition of non-B conformations by DNA at these motifs (e.g. hairpins, cruciforms, G-quadruplex, Z-DNA and H-DNA) as a common culprit through which they exert their biological effects possibly in concert with DNA metabolic and other DNA-related processes (4). Indeed, an increasing number of experiments mainly carried out in bacterial and yeast model systems indicates that long single DNA repeats [i.e. >150 bp; (3)] with the potential to form secondary structures (e.g. hairpins and cruciforms) can serve as targets for the shuffling and exchange of genetic information (2,3).
The knowledge about the biological activity of different types of DNA repeats in relation to gene repair pathways (especially single repeats in the size range <150 bp) and the putative role played in these processes by DNA replication is scant. This knowledge gap is particularly acute in cells of higher eukaryotes (2,3). In this study, we have devised an extrachromosomal functional read-out system based on pairs of complementary DNA templates carrying defective GFP-encoding sequences that can serve as substrates for intermolecular HR-dependent gene repair. This experimental system allowed us to investigate in a quantitative manner the effect of various types of single DNA repeats on the HR process in mammalian cells. Furthermore, by endowing acceptor DNA molecules with a eukaryotic origin of replication, we could probe in a strict manner the role of template DNA synthesis on repeat-induced homology-directed gene repair. We found that, in contrary to direct and spaced inverted repeats, both simple palindromes and composite inverted DNA repeats constitute targets for the HR pathway in mammalian cells. Induction of homology-directed gene repair was dependent on the arrangement and spacing of the repetitive DNA unit rather than on its nucleotide sequence. We also found that the presence of inverted DNA repeat sequences in target molecules rendered them susceptible to coordinated nicking by T7 endonuclease I, a bona fide four-way DNA branch resolving enzyme (29). These results are consistent with other in vitro data showing that lineform-to-cruciform transition in double-stranded DNA molecules relies on the presence of an inverted repeat and is negatively affected by intervening spacer sequences in a length-dependent manner (22–24). Thus, we demonstrate that nonspaced inverted DNA repeats per se can stimulate homology-directed gene repair in mammalian cells presumably due to their capacity to form secondary structures in vivo that can subsequently serve as direct targets for cellular structure-specific nucleases (Figure 4). These processes may eventually lead to the formation of DSBs that constitute canonical substrates for, amongst others, NHEJ- and HR-based allelic and non-allelic recombination. Indeed, Inagaki and coworkers have recently shown in 293 cells by using a two-plasmid system together with a PCR-based assay that large secondary structure-forming palindromic AT-rich repeats (PATRRs), often associated with translocations in the human germ line, stimulate intermolecular rearrangements via a pathway likely to involve NHEJ (27). The impact of template DNA replication on the PATRR-specific rearrangements was, however, not investigated.
Resolution of cruciform-like structures is thought to start with the introduction of single-strand breaks on opposite sites of the branch point followed by a ligation step resulting in the generation of hairpin-capped termini that can be further processed by nicking to generate “open” ends. Candidate resolving and processing enzymatic activities are those of the first isolated bona fide mammalian Holliday junction resolvase Gen1 (30) and Mre11 (31), respectively. Other candidate resolvase is that corresponding to the SLX4 complex (32). Possible outcomes of such ectopic recombination processes include chromosomal translocations and loss-of-heterozigosity. Related to this, in silico analysis of the human genome and experiments in yeast suggest that, during evolution, palindromes and inverted repeats with short spacers are counter selected compared with direct repeats and inverted repeats with long spacers (9). Indeed, without implying causality, more recent computer-aided phylogenetic sequence analyses revealed a correlation between DNA repeat pairs, NHEJ and non-allelic HR in the shaping of mammalian genome evolution (33).
Finally, we also showed that, at least under conditions that do not disrupt processivity of DNA synthesis, replication of molecules harboring the direct or the inverted DNA repeats did not significantly increase the frequencies of HR-dependent gene repair events when compared with those measured in the absence of acceptor DNA replication. This finding on single DNA repeats adds to recent results indicating that, at least in the case of the high-copy-number trinucleotide repeat associated with Friedreich’s ataxia GAA·TTC, DNA rearrangements can ensue in the absence of replication (34). Other processes like DNA transcription and certain repair pathways such as the herein examined HR can provide for alternative mechanisms underlying DNA repeat-associated rearrangements as proposed elsewhere (1,35). Indeed, the fact that repeat-associated DNA instability can occur independently of a replication-based mechanism (e.g. replication stalling or slippage) can also be circumstantially inferred from the significant age-dependent expansion of secondary structure-forming trinucleotide repeats in post-mitotic neurons of patients afflicted by certain neurodegenerative disorders (1).
Current models of inverted repeat-driven secondary structure formation in vivo posit that palindromes or quasi-palindromes can, under torsional strain, transit from line-form to cruciform in double-stranded DNA via intrastrand annealing. On the other hand, spaced inverted repeats, although being also self-complementary, can only hybridize in the single-stranded form such as when a DNA replication fork advances through them and concomitantly gives rise to the Okazaki initiation zone (OIZ) in the lagging strand. Possibly, under these conditions, and depending on the length of the repeat/spacer sequences relative to that of the OIZ, lagging strand self-annealing becomes thermodynamically favorable resulting in the formation of hairpins that can stall DNA replication (2,36). Interestingly, we showed that replication of DNA molecules containing the spaced inverted repeat spIR.1 could not overcome their inability to promote homology-directed gene repair. Related to this, Voineagu and colleagues (36) have recently deployed a SV40 ori-based plasmid system and 2D agarose gel electrophoresis to demonstrate that Alu-derived 320-bp-long inverted repeats with no spacer or with a relatively short 12-bp spacer lead to replisome stalling in COS-1 cells, whereas the same inverted repeat with a 52-bp spacer did not. The authors interpreted these results as a consequence of the OIZ size limit not allowing effective stem-loop hairpin formation by inverted DNA repeats with the larger spacer. Thus, on the basis of our results and those of Voineagu and coworkers and toward dissecting the inverted repeat parameters allowing hairpin assembly through the postulated lagging strand displacement-dependent mechanism, it will be interesting to evaluate different-sized repeat/spacer sequences and their relationship with replication fork stalling on one hand (36) and homology-directed gene repair on the other (this study). These experiments might help to define the rules underlying secondary structure formation, replisome stalling and the HR-inducing activity of inverted DNA repeats in mammalian cells. Finally, the functional genetic assay described herein might also be helpful in evaluating the effect of other types of DNA motifs/parameters on HR in higher eukaryotes and the contribution of cellular factors to this process.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials and Methods and Supplementary Figures 1–4.
FUNDING
Funding for open access charge: The research leading to these results has received funding from the European Community’s 7th Framework Programme for Research and Technological Development under grant agreement number 222878 (PERSIST).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Albert Pastink (Department of Toxicogenetics, Leiden University Medical Center) for critically reading the manuscript as well as Maria Jasin (Memorial Sloan-Kettering Cancer Center, New York, New York, USA) and Dongshen Duan (Department of Molecular Microbiology and Immunology, School of Medicine, University of Missouri, Columbia, Missouri, USA) for providing the I-SceI-encoding construct pCbASce and the AAV vector shuttle plasmid pDD2, respectively. We also thank New England Biolabs for making available cruciform-forming control plasmid pUC(AT).
REFERENCES
- 1.Pearson CE, Edamura KC, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 2.Lewis SM, Coté AG. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair. 2006;5:1146–1160. doi: 10.1016/j.dnarep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front. Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life. Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim EL, Peng H, Esparza FM, Maltchenko SZ, Stachowiak MK. Cruciform-extruding regulatory element controls cell-specific activity of the tyrosine hydroxylase gene promoter. Nucleic Acids Res. 1998;26:1793–1800. doi: 10.1093/nar/26.7.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber JE, Debatisse M. Gene amplification: yeast takes a turn. Cell. 2006;125:1237–1240. doi: 10.1016/j.cell.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Bikard D, Loot C, Baharoglu Z, Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010;74:570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eykelenboom JK, Blackwood JK, Okely E, Leach DRF. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, Van Ormondt H, Hoeben RC, van der Eb AJ. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 11.Gonçalves MAFV, van der Velde I, Janssen JM, Maassen BT, Heemskerk EH, Opstelten D-J, Knaän-Shanzer S, Valerio D, de Vries AAF. Efficient generation and amplification of high-capacity adeno-associated virus/adenovirus hybrid vectors. J. Virol. 2002;76:10734–10744. doi: 10.1128/JVI.76.21.10734-10744.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Nierop GP, de Vries AAF, Holkers M, Vrijsen KR, Gonçalves MAFV. Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease. Nucleic Acids Res. 2009;37:5725–5736. doi: 10.1093/nar/gkp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue Y, Dongsheng D. Development of multiple cloning site cis-vectors for recombinant adeno-associated virus production. Biotechniques. 2002;33:676–678. doi: 10.2144/02333dd03. [DOI] [PubMed] [Google Scholar]
- 14.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 15.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;313:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves MAFV, Pau MG, de Vries AAF, Valerio D. Generation of a high-capacity hybrid vector: packaging of recombinant adenoassociated virus replicative intermediates in adenovirus capsids overcomes the limited cloning capacity of adenoassociated virus vectors. Virology. 2001;288:236–246. doi: 10.1006/viro.2001.1073. [DOI] [PubMed] [Google Scholar]
- 19.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren J, Qu X, Chaires JB, Trempe JP, Dignam SS, Dignam JD. Spectral and physical characterization of the inverted terminal repeat DNA structure from adenoassociated virus 2. Nucleic Acids Res. 1999;27:1985–1990. doi: 10.1093/nar/27.9.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonçalves MAFV. Adeno-associated virus: from defective virus to effective vector. Virol. J. 2005;2:e43. doi: 10.1186/1743-422X-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vologodskii AV, Frank-Kamenetskii MD. The relaxation time for a cruciform structure in superhelical DNA. FEBS Lett. 1983;160:173–176. doi: 10.1016/0014-5793(83)80960-0. [DOI] [PubMed] [Google Scholar]
- 23.Sinden RR, Zheng GX, Brankamp RG, Allen KN. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics. 1991;129:991–1005. doi: 10.1093/genetics/129.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogo H, Inagaki H, Ohye T, Kato T, Emanuel BS, Kurahashi H. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato T, Inagaki H, Kogo H, Ohye T, Yamada K, Emanuel BS, Kurahashi H. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum. Mol. Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- 26.Cunningham LA, Coté AG, Cam-Ozdemir C, Lewis SM. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocations in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Declais A-C, Lilley DMJ. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opi. Struct. Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 31.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 32.Svendsen JM, Harper JW. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24:521–536. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Bourque G. Recovering genome rearrangements in the mammalian phylogeny. Genome Res. 2009;19:934–942. doi: 10.1101/gr.086009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA·TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Vasquez KM. Models for chromosomal replication-independent non-B DNA structure-induced genetic instability. Mol. Carcinogen. 2009;48:286–298. doi: 10.1002/mc.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.