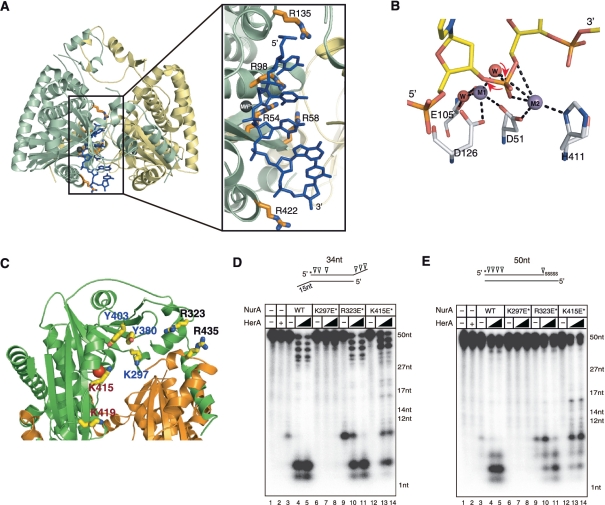
Figure 6.
PfNurA–ssDNA-binding model. (A) A ssDNA (blue) is modeled on PfNurA based on the TtAgo–DNA and PfNurA–dAMP–Mn2+ structures. Box: a close up view of the model showing the direction of the ssDNA, where the 3′ OH of a ribose ring points toward the open channel (entrance) and the 5′-phosphate group is directed to the narrow part of the channel. (B) Schematic drawing of the PfNurA nuclease mechanism. Mn2+ (M1) is coordinated by Asp51, Asp126 and two water molecules. Mn2+ (M2) interacts with Asp51, His411 and one water molecule. In this model, a water nucleophile (red) bound to both metal ions attacks the phosphate group and M1 stabilizes the leaving 3′ oxyanion, which allows the 5′–3′ digestion.(C) Structure of the PfNurA showing the position of some residues on the surface of the elliptical channel. Residues containing K297E/Y380F/Y403F (blue label), R323E/R435E (black label) and K415E/K419E (red label) are shown. (D) Nuclease activity assay of the three DNA-binding mutant using TP 424/423 or (E) TP 580/124. Each DNA substrate (20 nM) was incubated for 120 min at 65°C with 350 nM of PfNurA, increasing amounts of PfHerA (35 and 70 nM), 5 mM MnCl2 and 1 mM ATP. In lane 2, 70 nM of PfHerA was used as a control. Abbreviations: K297E*, K297E/Y380F/Y403F; R323E*, R323E/R435E; K415E*, K415E/K419E.