Figure 1.
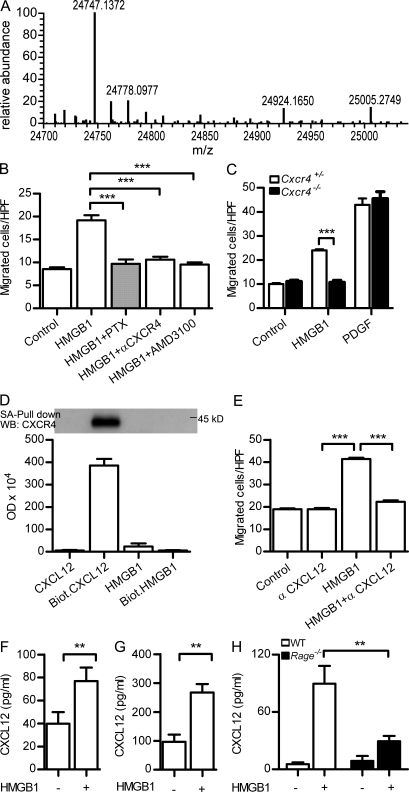
Migration of mouse fibroblasts in the presence of HMGB1 requires CXCR4/CXCL12. (A) Deconvoluted mass spectra of recombinant HMGB1 used throughout our work. The mass of 24,747 corresponds to the fully reduced state of HMGB1 (amino acids 2–215; the first methionine is absent both in mammalian and in bacterially produced HMGB1). (B) Migration of murine NIH/3T3 in the presence of 1 nM HMGB1 is 100 nM PTX–sensitive, and it is blocked by 2 µg/ml anti-CXCR4 antibody or by 1 µM of the CXCR4 antagonist AMD3100. Migrated cells were counted per high-power field (HPF) and are shown as mean ± SEM of three independent experiments (***, P < 0.005 vs. HMGB1 using ANOVA plus Dunnett’s test). (C) Migration of Cxcr4+/− and Cxcr4−/− embryonic fibroblasts (MEFs) in the presence of 1 nM HMGB1 or 10 ng/ml PDGF. Migrated cells were counted per high-power field and are shown as mean ± SEM of three independent experiments (***, P < 0.001 using ANOVA plus Bonferroni posttest). (D) Pull-down of CXCR4 from cell lysates of pre-B 300.19–CXCR4+ was performed with 500 nM of biotinylated CXCL12 or 500 nM of biotinylated HMGB1 and analyzed by Western blot. Nonbiotinylated CXCL12 or HMGB1 was used at 500 nM as control. Densitometric analysis of the bands obtained in three independent experiments (mean ± SEM) is shown. (E) Migration in the presence of HMGB1 is blocked by 1 µg/ml anti-CXCL12. Migrated cells were counted per high-power field and are shown as mean ± SEM of three independent experiments (***, P < 0.005 vs. HMGB1 using ANOVA plus Dunnett’s test). (F–H) CXCL12 as detected by ELISA in the supernatant of mouse 3T3 (F), human monocytes (G), or Rage−/− MEFs (H) stimulated with 4 nM HMGB1 for 2 h. Mean ± SEM of three independent experiments is shown (**, P < 0.01).