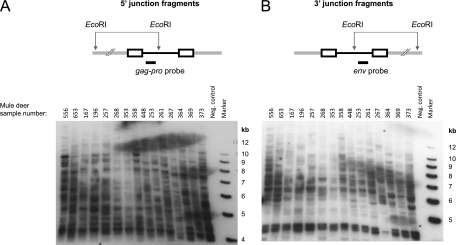
Fig 1.
Southern blot analysis of CrERVγ integrations in the mule deer genome. Mule deer genomic DNAs digested with EcoRI and transferred to a membrane were hybridized with a CrERVγ gag-pro probe to reveal the 5′ virus-host junction fragments (A) or a CrERVγ env probe to reveal the 3′ junction fragments (B). In the full-length CrERVγ genome, the distance of the EcoRI site to the 5′ and 3′ provirus ends is approximately 3.7 and 5.3 kb, respectively. The upper panels show schematic representations of the virus integration sites. The CrERVγ provirus is black, the virus LTR sequences are shown as rectangles, and mule deer flanking genomic DNA is shown as a gray double line. Size markers and mule deer identification numbers are indicated on the blots.