Fig 2.
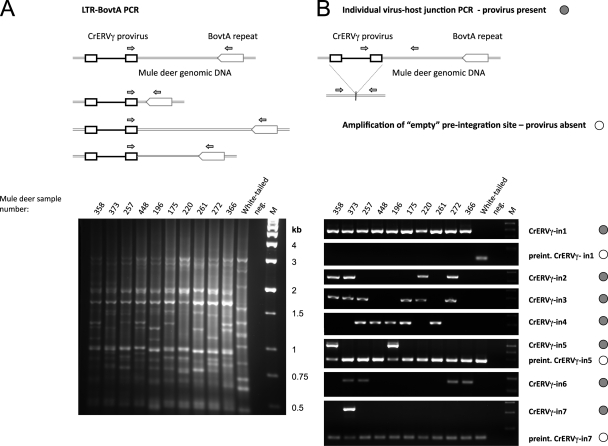
Polymorphic CrERVγ integrations detected by PCR of virus-host junction fragments. (A) Schematic representation and results of the LTR-BovtA PCR are shown. Primers, depicted as small arrows, were designed in a conserved region of the CrERVγ LTR and in a conserved part of the cervid BovtA genomic repeat. The PCR yields a pattern of bands, each representing a CrERVγ integration site with distinct distance to the nearest BovtA repeat. (B) PCR amplification of individual CrERVγ integration sites. The upper panels show schematics of the amplification of individual virus-host junctions, with primers (arrows) designed in internal virus sequences and in flanking mule deer genomic DNA away from the BovtA repeats. The 5′ and 3′ junctions were amplified; the 3′ junction scenario is depicted here. The amplification of the empty preintegration site is depicted schematically with primers in the 5′- and 3′-flanking regions relative to the virus integration point. The results of the PCR amplifications from mule deer and white-tailed deer genomic DNAs are shown below. The junction PCRs that detect the presence of specific virus integration are marked with filled circles, and the PCRs that amplify empty integration sites are marked with empty circles. For CrERVγ-in5 and CrERVγ-in7 the virus-host junctions and the empty integration sites were amplified in duplex PCRs with all primers present. M, molecular size marker; neg., no-template control; mule deer identification numbers are indicated on the gel.