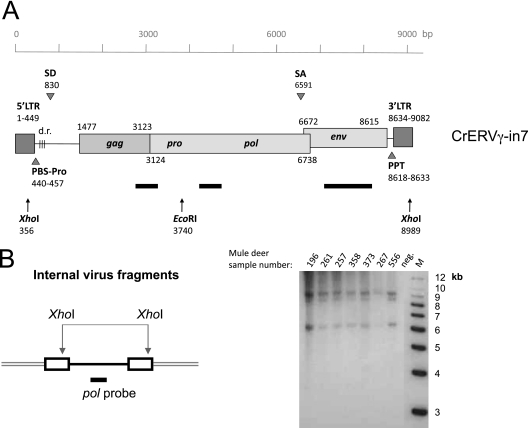
Fig 3.
Presence of full-length CrERVγ provirus in the mule deer genome. (A) Schematic diagram of full-length CrERVγ-in7 provirus. The scale references nucleotide positions, and the positions of predicted features are indicated (accession number JN592050). Black bars show positions of DNA probes used for Southern blot analysis, and arrows depict restriction enzyme recognition sites. SD, splice donor; SA, splice acceptor; PPT, polypurine tract; PBS, primer binding site; d.r., direct repeats. (B) Mule deer genomic DNAs digested with XhoI and hybridized with a CrERVγ pol probe to reveal the length of complete virus fragments. Three viral species can be detected in animals tested, representing full-length genomes and two with deletions. M, molecular size markers; neg., negative control (no DNA). Mule deer identification numbers are indicated on the gel.