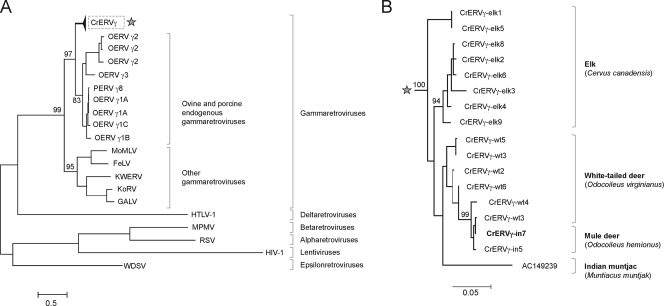
Fig 4.
Phylogenetic relatedness of CrERVγ. (A) A maximum-likelihood tree based on a 0.9-kb fragment of retroviral pro/pol nucleotide sequence obtained from GenBank and sequences obtained in this study for CrERVγ shows the placement of CrERVγ in the gammaretroviruses. (B) A branch of the tree from panel A (highlighted by an asterisk) representing the CrERVγ sequences from cervid species is shown in expanded form. The same region of pro/pol as that used for panel A was amplified by PCR from elk and white-tailed deer. The Indian muntjac sequence was obtained from GenBank. Bootstrap support is shown at branch nodes. The scale bars refer to the number of substitutions per site. Shown are PERV γ8 (porcine endogenous retrovirus; GenBank accession no. AF511112), OERV (ovine endogenous retrovirus; accession numbers AY193896 through AY193903), MoMLV (Moloney murine leukemia virus; NC_001501), FeLV (feline leukemia virus; NC_001940), GALV (gibbon ape leukemia virus, NC_001885); KoRV (koala retrovirus, AF151794); KWERV (killer whale endogenous retrovirus, GQ222416), HIV-1 (K03455), WDSV (walleye dermal sarcoma virus; AF033822), HTLV-1 (human T-cell lymphotropic virus; D13784), MPMV (Mason-Pfizer monkey virus; NC_001550), and RSV (Rous sarcoma virus; AF033808).