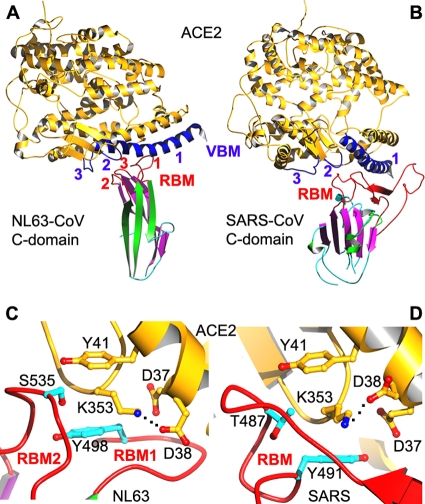
Fig 3.
Receptor binding by alpha-coronavirus NL63-CoV and beta-coronavirus SARS-CoV C-domains. (A) Crystal structure of NL63-CoV C-domain complexed with human ACE2 (PDB identification no. 3KBH). (B) Crystal structure of SARS-CoV C-domain complexed with human ACE2 (PDB identification no. 2AJF). (C) A hydrophobic tunnel structure at the NL63-CoV/ACE2 interface, comprising residues Tyr41 and Asp37 from ACE2 and Ser535 and Tyr498 from the NL63-CoV C-domain. (D) A hydrophobic tunnel structure at the SARS-CoV/ACE2 interface, comprising residues Tyr41 and Asp37 from ACE2 and Thr487 and Tyr491 from the SARS-CoV C-domain. The hydrophobic tunnels in panels C and D bury a salt bridge between Lys353 and Asp38 from ACE2. ACE2, angiotensin-converting enzyme 2; VBM, virus-binding motif.