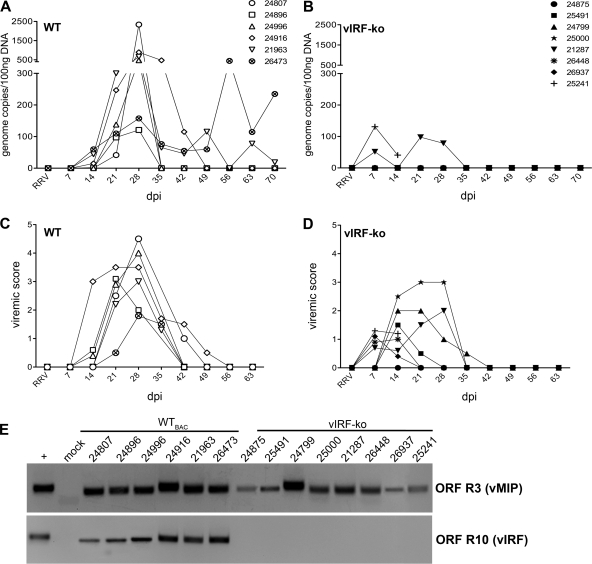
Fig 1.
Detection of RRV DNA and lytic replication measured in RRV-infected RMs. Expanded-specific-pathogen-free RMs were infected intravenously with 5 × 106 PFU of WTBAC RRV (n = 6) or vIRF-ko RRV (n = 8). Each RM is labeled with an identifying number and an individual symbol. (A and B) Whole blood was analyzed weekly via qPCR to determine RRV loads. Data are represented as number of RRV genome copies/100 ng DNA in each of the WTBAC RRV-infected RMs (A) and the vIRF-ko RRV-infected RMs (B). (C and D) PBMCs (2 × 105) were cocultured with confluent RFs and serially diluted (1:3) across a 24-well plate. Viremic score correlates with the highest dilution of PBMCs that resulted in CPE when cocultured with RFs. A viremic score of 5 indicates the presence of CPE in wells with the highest dilution of PBMCs (2.5 × 103 PBMCs), and the presence of CPE in duplicate wells at each dilution has a viremic score of 0.5. (E) The presence of RRV DNA in cocultures was verified via nested PCR using primers for RRV ORF R3 (vMIP), as well as RRV ORF R10 (vIRF), to differentiate WT and vIRF-ko RRV.