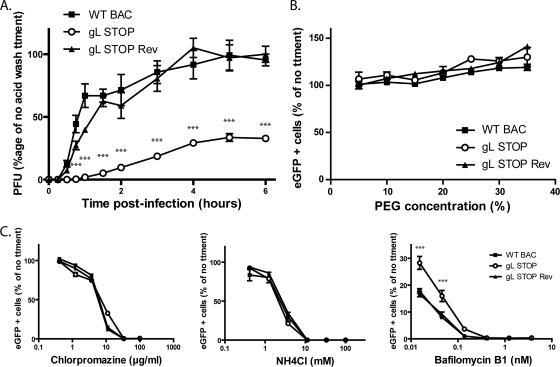
Fig 6.
The entry deficit of the BoHV-4 gL STOP strain is linked with a defect in penetration. (A) MDBK cells were exposed to eGFP-expressing wild-type (WT BAC), gL STOP, or gL STOP Rev BoHV-4 strains (200 PFU/well) for 2 h on ice to allow virus binding but block virus penetration before being washed three times with ice-cold PBS (pH 7.4) to remove unbound virions. After incubation at 37°C for the indicated times, the cells were washed with isotonic acid (pH 3) buffer and overlaid with complete medium containing 0.6% medium-viscosity carboxymethylcellulose (Sigma). At 96 h postinfection, viral plaques (eGFP positive) were counted. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests (***, P < 0.001). (B) PEG-induced fusion of gL− virions. Cells were infected with eGFP-expressing WT BAC, gL STOP, or gL STOP Rev BoHV-4 strains (37°C, 0.5 PFU/cell). After 2 h, the cells were washed in PBS, and membrane fusion of cell surface virions was induced by PEG treatment at different concentrations. Eighteen hours after the virus was added to the cells, viral infection was assayed by flow cytometry for eGFP expression. Results are expressed as percentages of the levels seen with no PEG treatment. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests. No statistical difference was observed between viral strains. (C) Dependence of WT and gL-deficient virions on endocytosis and lysosomal acidification. MDBK cells were incubated for 1 h with drugs as shown and then exposed to eGFP+ WT BAC, gL STOP, or gL STOP Rev BoHV-4 virions (0.5 PFU per cell). Drug treatment was maintained throughout the course of infection. Infection rates were determined by a flow cytometric assay of viral eGFP expression after 18 h. The data presented are the averages of triplicate measurements ± standard errors of the means and were analyzed by two-way ANOVA and Bonferroni posttests (***, P < 0.001).