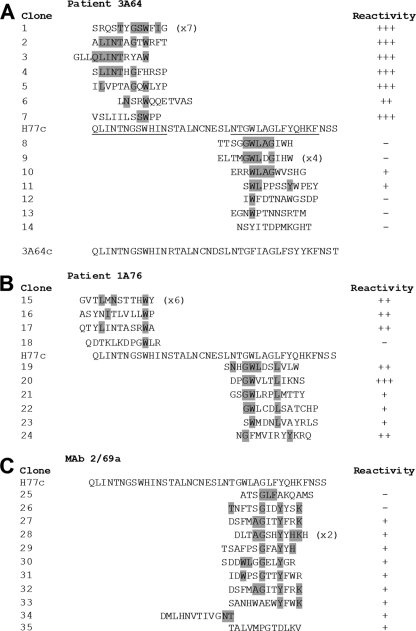
Fig 8.
Epitope mapping by enrichment of random peptide phage display libraries reveals the presence of distinct yet overlapping epitopes within regions of E2 encompassing aa 412 to 423 and aa 434 to 446. Affinity-purified Ig from patients 3A64 (A) and 1A76 (B) and MAb 2/69a (C) were used to enrich phages from a 12-mer random peptide library. Following three to four rounds of biopanning, individual phage clones were isolated, amplified, and tested for reactivity to the selecting antibody, and the amino acid sequence of the peptide insert was determined by DNA sequencing. The resulting sequences were aligned to the corresponding region of the H77c E2 amino acid sequence. The phage clone identification is presented, and individual residues aligning to the H77c sequence are shaded. Numbers in parentheses indicate the number of times each peptide sequence was recovered. The reactivity of each phage to the selecting antibody in a phage capture enzyme immunoassay is shown alongside each peptide sequence and is shown as the relative optical density compared to that of capture by an antibody preparation from an HCV-negative donor (NHS-Ig). +++, >4-fold greater than the optical density obtained for the negative control NHS-IgG; ++ >3-fold greater; +, >1.5-fold greater; −, <1.5-fold greater. In addition, the consensuses of the RNA sequences of the viruses infecting patient 3A64 were deduced by reverse transcription-PCR (RT-PCR) and sequencing (A). This sequence is presented below the alignment for the peptides selected by antibodies isolated from patient 3A64.