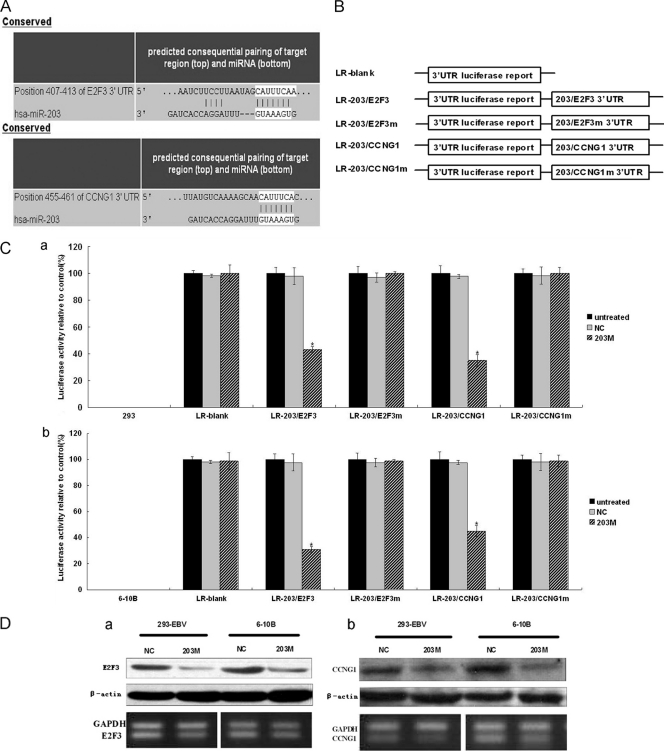
Fig 3.
Identification of the target genes of miR-203 and the effect of miR-203 mimics on those targets. (A) Base pairing between miR-203 and the target genes, E2F3 and CCNG1, was predicted by TargetScan software. (B) Schematic representation of the five reporter constructs, named LR-blank, LR-203/E2F3, LR-203/E2F3m, LR-203/CCNG1, and LR-203/CCNG1m, respectively, where “m” indicates the mutant construct with 5 or 6 bases deleted at the predicted binding site of the gene 3′ UTRs. LR, luciferase reporter. (C) Each of the constructed luciferase reporter plasmids was cotransfected with the indicated material (untreated; NC, scramble; 203 M, miR-203 mimic) into the 293 (a) or 6-10B NPC (b) cells. Luciferase activities were measured after 48 h, and β-galactosidase was used for normalization of the difference of transfection efficiencies. The result showed that miR-203 combined with the predicted sites at the 3′ UTRs of the two genes, resulting in decreased luciferase activity. Data were confirmed in three experiments. A P value of <0.05 (*) was considered to be statistically significant. (D) The effect of miR-203 mimics on the expression of endogenous E2F3 and cyclin G1. The 293-EBV cell line or the NPC-derived cell line, 6-10B, was treated with 203 M or NC; after 48 h of the treatment, E2F3 (a) and cyclin G1 (b) at both protein and mRNA levels were assayed by Western blotting (upper panels) or semiquantitative RT-PCR (lower panels) analysis.