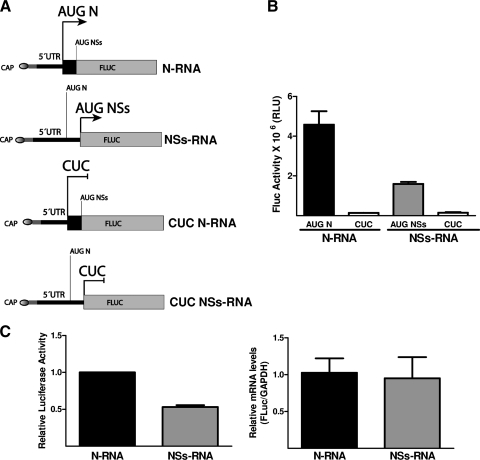
Fig 3.
NSs ORF can be translated in the context of virus-like smRNAs. (A) Schematic representation of the capped monocistronic RNA constructs that mimic the ANDV S mRNA. To faithfully mimic the viral mRNA, these RNAs contain 21 additional nucleotides (5′-GGGAGACCCAAGCTGGCTAGC-3′; gray region) between the 5′ cap structure (oval at the 5′ end of the RNA) and the first nucleotide of the ANDV smRNA sequence (black line) (50). Additionally, the viral NSs ORF has been replaced by the firefly luciferase (FLuc) reporter gene. The arrow indicates the initiation codon used. In the N RNA, the N initiation codon is in frame with FLuc. In the NSs RNA, the putative NSs initiation codon is in frame with FLuc. In the CUC RNAs, the AUGN or AUGNSs codons have been mutated to CUC. (B) The N RNA (black bar) and NSs RNA (gray bar) were translated in a commercial in vitro translation system as indicated in Materials and Methods. FLuc activity (in relative light units [RLU]) was measured as indicated in Materials and Methods. Unshaded bars correspond to FLuc activity obtained with CUC mutations. Values are the means ± standard deviations (SD) from three independent experiments, each conducted in triplicate. (C) DNA plasmids coding for the RNAs depicted in panel A plus an RLuc-expressing plasmid (transfection efficiency control) were transfected into HeLa cells. Total RNA and protein were extracted and analyzed. RNA was used to assess the total amount of the virus-like RNAs in cells by an RT-qPCR assay (right panel). The quantity of N RNA was arbitrarily set to 1, and the total amount of NSs RNA recovered from cells was expressed relative to this value (right panel). FLuc and RLuc activities were determined, and FLuc activity was normalized to RLuc activity (left panel). Normalized N RNA FLuc activity was arbitrarily set to 1 (left panel). Values shown in both panels are the means ± SD from three independent experiments each conducted in triplicate.