Abstract
Phage display libraries are widely used as tools for identifying, dissecting and optimizing ligands. Development of a simple method to access greater library diversities could expedite and expand the technique. This paper reports progress toward harnessing the naturally occurring diversity generating retroelement used by Bordetella bronchiseptica bacteriophage to alter its tail-fiber protein. Mutagenesis and testing identified four sites amenable to the insertion of <19-residue heterologous peptides within the variable region. Such sites allow auto-generation of peptide libraries surrounded by a scaffold with additional variations. The resultant self-made phage libraries were used successfully for selections targeting anti-FLAG antibody, immobilized metal affinity chromatography microtiter plates and HIV-1 gp41. The reported experiments demonstrate the utility of the major tropism determinant protein of B.bronchiseptica as a natural scaffold for diverse, phage-constructed libraries with heterologous self-made phage libraries.
Keywords: diversity generating retroelement, mutagenesis, phage display, protein engineering, selections
Introduction
Molecular display provides a powerful tool for binding partner identification and optimization of receptor and biopharmaceutical binding (Scott and Smith, 1990; Atwell et al., 1997; Pedersen et al., 1998; Sidhu, 2000; Smothers et al., 2002; Doyon et al., 2003; Kehoe and Kay, 2005). For example, the immune system is a model of a naturally occurring molecular display library for the recognition of diverse binding partners. Such capabilities are approached by the best molecular display systems used in laboratories with maximum diversities of 1014 different protein variants per library (Hanes and Plückthun, 1997; Daugherty et al., 1999; Wilson et al., 2001; Yeung and Wittrup, 2002; Diaz et al., 2003). Improvements to protein diversity generation and optimization could enable tremendous practical benefits. This study aims to harness the diversity generating retroelement (DGR) used by Bordetella bacteriophage (BP) to generate 1013 tail-fiber variants.
The naturally occurring self-made phage library (SMPL) used by bacteriophage infecting Bordetella bronchiseptica could offer vast diversity in a more expedient format than conventional molecular display systems. The wild-type BP SMPL auto-generates ∼1013 different protein sequences (Liu et al., 2002). This diversity exceeds conventional phage display methods and the theoretical 1011 diversities accessed by antibody rearrangement in the immune response (Nemazee, 2006). Among molecular display systems, we are unaware of any comparable approach for the self-generation of protein libraries.
Combinatorial libraries of tail-fiber proteins provide an advantage to BP when binding to its host. BP, a T7-type phage, attaches to host cell surface receptors before injecting its DNA into the cell. After integration of phage DNA into the host's genome, the phage becomes lytic only in the presence of environmental cues, such as UV irradiation or rapid growth of the host. The BP host bacteria can cause respiratory infections in humans and small mammals, and transitions between virulent (Bvg+) and avirulent (Bvg−) phases of its life cycle (Akerley et al., 1995; Uhl and Miller, 1996). During this transition, the outer bacterial surface acquires different cell surface proteins (Akerley and Miller, 1996). To maintain its infectivity, BP must change its tail-fiber protein to adhere to a dynamic array of host-displayed receptors. The adaptive diversity generating capability gives BP a selective advantage for infecting its host, and provides an intriguing system to exploit for the engineering of new proteins.
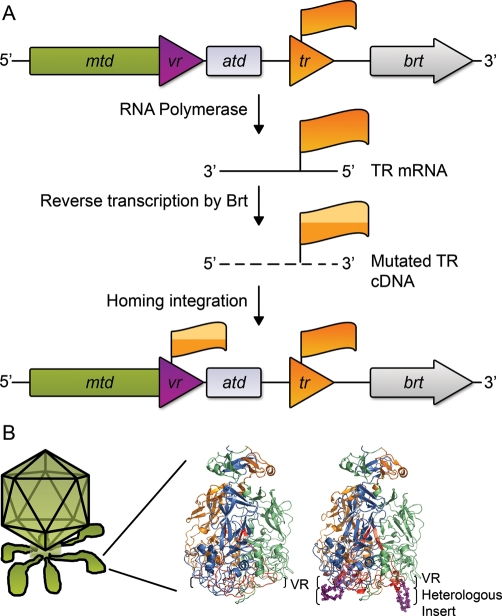
A DGR synthesizes the BP SMPL (Fig. 1A). This DGR mutates the major tropism determinant (mtd) gene encoding a phage tail-fiber protein (Fig. 1B). The resultant SMPL displayed on the tail fiber allows host infection despite changes to the composition of the host surface. The DGR introduces randomized DNA sequences into specific sites of the variable region (vr) of the mtd open reading frame. The randomized sequences are cDNA transcribed from the non-coding template region (tr) (Fig. 1A). By directing the mutagenesis to the residues responsible for phage–host interactions, the DGR avoids disrupting the trimeric fold of the Mtd, and only alters the solvent-exposed variable region (VR), which is encoded by the vr, the C-terminal part of the mtd gene (Liu, et al., 2002; Doulatov et al., 2004; Liu et al., 2004; McMahon et al., 2005; Dai et al., 2010) (Fig. 1B).
Fig. 1.
The SMPL system for molecular display. (A) Testing the diversity generating mechanism used by the BP SMPL system. In this report, heterologous sequences (solid orange flag) encoding peptide tags were inserted into the template region or non-protein coding tr. The tr mRNA is reverse-transcribed by Brt in a putatively adenine error-prone manner. The mutated tr cDNA transcript is then integrated into the vr region of the mtd. (B) The Mtd trimer on the tail fibers of a schematic BP. The crystal structure of the Mtd trimer (center, PDB coordinates 1YU4) accommodates the expression of 1013 peptide variants across the VR. A homology model illustrates heterologous display of 14 amino acids C-terminal to Mtd position 367 (purple). The insert and protein structure figures throughout the paper were prepared using ICM Pro and Pymol (Abagyan et al., 1997; DeLano and Lam, 2005).
The DGR requires six known components (numbered in bold font below). First, the mtd encodes two bell-shaped trimeric proteins (Mtd) attached to each of the six phage tail fibers yielding 36 copies of the Mtd per phage (Fig. 1B) (Dai et al., 2010). The Mtd forms a C-type lectin fold, and provides a structured scaffold for the display of the VR, the second component of the DGR (McMahon et al., 2005). At the C-terminus of the Mtd, the VR forms the base of the tail-fiber proteins, which interacts with the host. The 134 base-pair (bp) vr, encodes the VR, and, like the variable region of antibodies, undergoes a high rate of mutagenesis. The mutagenesis of the vr occurs at 12 variable codons. Translation of the vr can result in a library with up to 1013 different VR variants displayed on the phage tail fibers (Supplementary Fig. S1).
The third and fourth of six DGR components, the 134 bp non-protein encoding template region (tr) and phage encoded reverse transcriptase, Brt, are the basis for the SMPL generation. The tr, an invariant copy of the vr, contains 12 adenine-containing, mutation-prone codons that correspond to the 12 variable codons of the vr. The putatively error-prone Brt generates sequence variability during reverse transcription of the tr mRNA into mutant cDNA. This key step diversifies the tr mRNA by replacing adenine bases of the mutation-prone codons with any of the four DNA bases before integration into the vr to create the SMPL (Fig. 1A). Mutation of one or both adenines of the mutation-prone codons can encode either 4 or 15 different amino acids depending upon the codon altered; the system avoids introducing stop codons as adenines appear in only the first or second positions of the 12 targeted codons. Fifth, the initiator of mutagenic homing (imh), a 21 bp sequence located at the 3′ end of both the tr and vr sequences, controls the directionality of tr to vr sequence transfer, termed ‘mutagenic homing’ (Doulatov et al., 2004). Sixth, the Atd has an unknown, but necessary, function and likely assists with mutagenic homing (Guo et al., 2008). The six components of the DGR provide the known requirements for both adenine-specific mutagenesis and potentially protein engineering (Fig. 1).
Previously, we used an SMPL of Mtd protein variants to identify binding partners to T4 lysozyme. After expression as a GST fusion protein, Mtd selectants from the SMPL bound T4 lysozyme with high affinity and specificity (Yuan and Weiss, Unpublished data). Here, we hypothesize that a heterologous DNA sequence introduced into the tr could also undergo adenine-targeted mutagenesis (Fig. 1A). When transferred to the vr, the Mtd would display a library of heterologous peptides as a fusion to the phage tail fiber (Figs 1B and 2). The diversity of the resultant heterologous library would be limited only by the number of adenines encoded in the insert DNA (Fig. 3), the successful transfer of the tr to vr and the requirements for phage propagation. Such SMPLs could generate library diversities constrained only by the culture volume. Here, we demonstrate successful selections with modified Mtd variants (Fig. 4). With exceptionally high mutation rates in a prokaryotic host, the BP SMPL could provide a powerful technology for protein engineering.
Fig. 2.
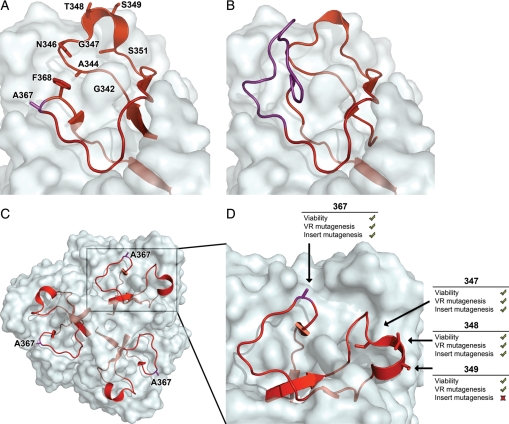
The VR display challenge. (A) Structure of the Mtd monomer with the 9 VR positions tested for heterologous display labeled. This view highlights one subunit of the trimer shown in Fig. 1. The position C-terminal to residue 367 (purple) can escape the deep canyon engulfing the VR, and allows insertion of a heterologous peptide without disruption of the Mtd trimer or phage–host interactions. (B) Surface diagram of the homology model of the 14 amino acid insertion (purple) C-terminal to position 367. (C) The base of the trimeric Mtd tail fiber showing the engulfed VR with position 367 highlighted. (D) Enlarged region of Mtd monomer highlighting four Mtd positions accepting peptide inserts without disruption of the DGR or phage infectivity.
Fig. 3.
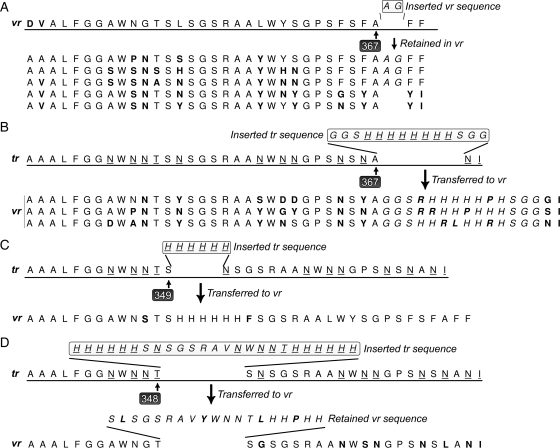
SMPL mutagenesis with test sequences. Mutated codons are in bold, insert codons are italicized and adenine containing codons of the tr are underlined. (A) To determine the viability of an insert C-terminal to Mtd position 367, a heterologous, non-adenine insert encoding a restriction site was constructed. A silent mutation (GCG→GCT) was also inserted at position 367 as a non-adenine marker for transfer. The tropism switching mechanism repaired the silent mutation and excised the restriction site. (B) To test a larger insert in position 367, a 14 codon sequence was inserted into position 367 and the entire insert was transferred to the VR. Both heterologous and endogenous adenine containing amino acids were mutated before insertion into VR, as shown. (C) The insertion of a His6 peptide at position 349 tested this region of the Mtd for both variability and viability. Seventeen percent of endogenous adenine containing amino acids were mutated; however, no heterologous mutagenesis occurred. (D) To test the ability of the DGR to mutate heterologous adenines in position 348, a 25 amino acid sequence was inserted into the tr. Nineteen codons of the insert were transferred in-frame to the vr with mutagenesis throughout the insert and surrounding scaffold.
Fig. 4.
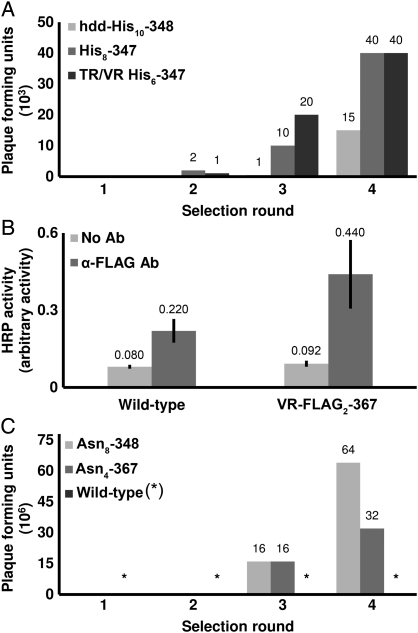
Selections with an SMPL. (A) Immobilized metal affinity chromatography microtiter plates were used as the capture target for three His-tagged epitope variants. (B) SMPL display. A BP variant displaying a FLAG epitope (Mtd FLAG2-367) or BP (wild type) were incubated in anti-FLAG-coated microtiter wells or control wells lacking the antibody. Error bars represent standard error (n = 3). (C) Selections for HIV gp41 binding. Target (gp41)-coated microtiter plates were used to capture mutants displaying a library based upon the sequences (AAC)4, (AAC)8 or wild-type BP (no insert) during four rounds of selections. Bound BP or mutants were eluted and amplified as in (A). Strong enrichment was observed for the SMPLs with the peptides inserted into the Mtd, and no enrichment resulted from the wild-type Mtd SMPL.
Materials and methods
Mutagenesis using phagemid templates and homologous recombination by tri-parental mating
Conventional, oligonucleotide-directed mutagenesis of sequences in the M13 phagemid was used to insert sequences into the tr and, for some inserts, the vr. This mutagenesis operated on the BP sequences flanked by 500 bp required for later homologous recombination into the BP genome (Kunkel, 1985; Sidhu et al., 2004). Briefly, uracil-doped phagemids were produced by growth in Escherichia coli CJ236 cells. The phagemids were then annealed to phosphorylated oligonucleotides encoding the desired mutations for insertion of the mutagenized heterologous sequences (Supplementary materials). After sequencing to verify mutagenesis, the mutant tr were subcloned into a suicide vector (pRE112) before transfer into B.bronchiseptica strain 6401 and homologous recombination into the BP genome as previously described (Figurski and Helinski, 1979; Edwards et al., 1998). Conventional polymerase chain reaction (PCR) protocols were used to amplify colonies from this tri-parental mating step followed by standard DNA sequencing methods to identify successful transformants. Colonies were then cultured and the supernatant filtered (0.2 µm pore filter) to obtain mutant phage. The soft-overlay method (Adams, 1959) was used to prepare the phage for viability and variability assays and to isolate tropism switching phage as previously described (Liu, et al., 2002). DNA from infective phage was sequenced according to standard protocols.
Selections and screens using immobilized metal affinity chromatography or target proteins
Phage variants isolated as described above were diluted 1 : 1 in dilution buffer [0.1% Tween 20 with 0.2% casein in phosphate-buffered saline (PBS)]. The targets were immobilized on microtiter plates. The His-tag library targeted His-Select HS Nickel-coated microtiter plates. The selections targeting anti-FLAG or HIV-1 gp-41 used Maxisorp immunoplates coated with the protein (10 mg/ml) in 50 mM sodium carbonate (pH 9.6), blocked with 0.2% non-fat milk. Then, 200 µl of ∼1012 filtered phage was added to the microtiter plates before incubation for 1 h on a rotary shaker at room temperature or at 4°C overnight. The plates were then washed 3–11 times with PT buffer (0.05% Tween 20 in PBS) before eluting bound phage with 100 µl 0.1 M imidazole for the His-tagged libraries or through addition of 100 µl 0.1 M HCl. The eluted phage was collected, and the solution was neutralized, if necessary, by addition of one-third volume 1 M Tris. Following titers, the phage were amplified for subsequent rounds of selections.
Phage-based enzyme-linked immunosorbent assays (ELISA) were used to examine phage affinity for target binding. A representative protocol for screening phage binding to the anti-FLAG antibody is provided here. Briefly, Maxisorp immunoplates (96 well) were coated with anti-FLAG antibody (1 : 1000) diluted in 50 mM sodium carbonate (pH 9.6), blocked with 0.2% non-fat milk and then washed with wash buffer (PBS supplemented with 0.05% Tween-20). A phage mutant with the FLAG epitope inserted as described or the wild-type phage was transferred to the antibody-coated plate. After a 1 h incubation, the plates were washed before incubation with rat-derived anti-Mtd antibody followed by three wash steps and the addition of anti-rat IgG conjugated to horseradish peroxidase. Next, the plates were developed using a solution of o-phenylenediamine dihydrochloride, and read spectrophotometrically at 450 nm with a 96-well microtiter plate reader.
Homology modeling using ICM Pro and PyMol
Models of Mtd trimers with the heterologous inserts were constructed using the ICM-Pro homology functions (Abagyan et al., 1997; DeLano and Lam, 2005) and the 1YU4 structure from the protein data bank (Figs 1B and 2B).
Results and discussion
Strategy for the identification of positions tolerating heterologous sequences
Harnessing the BP SMPL for foreign sequence mutagenesis requires the identification of one or more positions within the VR of Mtd that can tolerate the insertion of new peptides. Heterologous sequences inserted at the chosen position must not disrupt the Mtd trimer or phage interactions with its host. Furthermore, the DNA encoding the sequence must be amenable to the SMPL mutagenesis mechanism. Crystal structures (McMahon et al., 2005) and secondary structure predictions (Cheng et al., 2005) suggested nine positions, that could accommodate heterologous peptide insertion and display (Fig. 2A). The potentially amenable positions were targeted by adenine containing inserts encoding sequences within the tr and, for some inserts, the vr.
The design of >50 constructs to establish viable positions for heterologous inserts included different epitopes and locations within the vr and tr. The five tested epitopes included His-tag, FLAG, c-Myc, poly-lysine (AAA)8 and poly-asparagine (AAC)4 (Table I). Linkers between the inserted sequence and the Mtd included both flexible Gly–Ser sequences and a homodimerization domain known to form a stem-like structure (Gururaja et al., 2000; Marks et al., 2004), which could extend from the Mtd surface. Additionally, the linkers included a range of different lengths. Initially, each of the nine positions was tested for tolerance to an insert using five variations of the His-tag epitope to assess the availability of different configurations for binding (Table I). From the constructs designed, trends emerged for the phage infectivity, insert stability and SMPL formation capability.
Table I.
Epitope variants and insert positions tested
| Mtd insert position | Description | Sequence |
|---|---|---|
| TR VARIANTS | ||
| 342, 344, 346, 347, 348,349, 351, 367, 368 | His6 | HHHHHH |
| 342, 344, 346, 347, 348, 349, 351,367, 368 | His8 | GGSHHHHHHHHSGG |
| 348 | hdd-His10 | KFLIVKSGPAGHHHHHHHHHHGGAPGEFLIVES |
| 348 | His6LOOP | GGSGHHHHHHNGPHHHHHHGSGG |
| 348 | 2XHis | HHHHHHSNSGSRAVNWNNTHHHHHH |
| 342, 344, 346, 347, 348, 349, 351, 367, 368 | FLAG | LGDYKDDDDK |
| 367 | FLAG2 | GGSDYKDDDDKDYKDDDDKSGG |
| 348 | hdd-FLAG | KFLIVKSGPAGDYKDDDDKGGAPGEFLIVES |
| 367 | c-myc | GGSEQKLISEEDLSGG |
| 348, 367 | Lys8 | GGSKKKKKKKKSGG |
| 348,367 | Asn4 | GGSNNNNSGG |
| 348, 367 | Asn8 | GGSNNNNNNNNSGG |
| VR VARIANTS | ||
| 348, 368 | His6LOOP | GGSGHHHHHHNGPHHHHHHGSGG |
| 367 | His6 | HHHHHH |
| 367 | c-myc | GGSEQKLISEEDLSGG |
| 367 | Asn4 | GGSNNNNSGG |
| 367 | His8 | GGSHHHHHHHHSGG |
| 367 | FLAG2 | GGSDYKDDDDKDYKDDDDKSGG |
| 367 | AG | AG |
| TR/VR VARIANTS | ||
| 347 | His6 | HHHHHH |
| 348 | 2XHis/His6LOOP | HHHHHHSNSGSRAVNWNNTHHHHHH/GGSGHHHHHHNGPHHHHHHGSGG |
Results with underlined constructs are described in the text.
Residues not underlined failed viability or variability assays.
hdd, homodimerization domain (Marks et al., 2004).
Testing each site for tolerance to heterologous inserts required multiple steps. The inserts were introduced by the Kunkel method of oligonucleotide-directed mutagenesis targeting DNA within a phagemid (Kunkel, 1985). The mutated sequences were then subcloned into a suicide vector for transfer to Bvg+ bacteria via triparental mating (Materials and methods). This transfer step resulted in mutant prophage (Edwards et al., 1998; Stibitz, 1998; Cotter and Miller, 2001). Propagation to release the prophage then allowed testing for viability, as defined by infectivity of Bordetella bronchseptica. If the modified phage remained viable, further testing examined the ability to mutate variable codons of the tr and to detect transfer of heterologous inserts from tr to the mtd vr. DNA sequencing of the resultant phage tracked this mutagenesis; as expected (Guo et al., 2008) some of the sequenced clones were wild-type Mtd. Furthermore, DNA sequencing was sometimes complicated by multiple clones present in each plaque.
Heterologous inserts placed in the tr
The heterologous sequences inserted into the tr explored four possible outcomes. First, the insert could result in non-viable phage, which would be incapable of infecting B.bronchiseptica. Second, the insert could remain stably incorporated in the tr, but fail to transfer to the vr. Third, the insert could transfer from tr to vr but fail to undergo mutagenesis. Fourth and most desirable, the inserted sequences could transfer from tr to vr, undergo mutagenesis and result in viable phage capable of leveraging the SMPL mutagenesis.
Four of the nine tested positions (347, 348, 349 and 367) emerged as both viable and variable to tolerate insertions into the tr, the desirable fourth scenario above (Fig. 3, Table I). The five failed positions (342, 344, 346, 351 and 368) may result from peptides that were insufficiently surface exposed to allow transfer and remain viable (scenario two above) (Fig. 2A, Table I). After phage propagation of variants of the four successful positions, insert stability was then verified by PCR of plaques from B.bronchseptica infected phage. The subcloned inserts remained intact in the tr after one or more rounds of infection. The adenine-specific mutagenesis was observed in the variable codons of the wild-type endogenous vr for all four tr inserts. However, under current culture and detection methods, the heterologous insert only rarely transferred from tr to vr. Furthermore, the tr to vr transfer of the heterologous insert sometimes took place, and only the variable bases of the endogenous vr were altered. Taken together, the results indicate that the DGR mechanism can function as expected, despite new, exogenous bases inserted into the tr (Fig. 3A–D).
Testing insertion of peptides in Mtd position 367
Based on the successful insertion and transfer of heterologous DNA sequences described above, position 367 was explored further. Inserts at position 367 were chosen for experiments aimed at insertion of a short two-residue peptide into the VR (Fig. 3A). VR position 367 produced variant phage that remain infective, and have a functional DGR. Residue 367 provides a surface-exposed knuckle arcing away from a deep canyon wedged tightly around the VR. This exposure could allow the inserted peptide to extend away from the surrounding residues. Thus, inserts at this location avoid disrupting interactions with the host. The resultant phages were both variable and viable. Mutation-prone codons in the VR show extensive variability in five unique sequences (n = 19 sequenced including wild-type and mixed clones) providing further evidence that inserts at this position will not hinder the DGR, nor phage infectivity (Fig. 3A).
To further investigate the tolerance of Mtd 367 for a larger insert, GGSHis8SGG, a 14 amino acid insert was subcloned into the tr. The first series of inserts tested the compatibility for sequences encoding His8 peptides placed in the tr. This series also tests transfer of new sequences from tr to vr. The codons for histidine (CAT or CAC) include a mutable adenine for tracing mutagenesis during transfer. Transfer of the entire 42 bp insert from tr to vr was observed to initiate at the 3′ end of the vr with adenine-specific mutagenesis of the heterologous insert and throughout the length of the vr (Figs 2B and 3B). The histidine codons in the insert were mutated in three unique sequences (n = 5) to arginine, proline or leucine as expected from the BP DGR mutagenesis mechanism (Fig. 3B).
The mutation rate for the exogenous insert was 33%, whereas the mutation rate for the endogenous scaffold was as high as 83%. Thus, the construct generates a library of 6.6 × 104 peptide variants nested within the naturally occurring Mtd library of ∼9 × 1012 variants. This yields a new combinatorial library with a theoretical diversity equal to the product of the individual library diversities (5.9 × 1017 different potential variants). The practical diversity of this library is limited by only three constraints: the number of adenines contained within the heterologous DNA insert, the scale of bacteriophage cultured (which currently approaches 2 × 1015 phage per liter) and the rate of transfer from tr to vr. Each new generation of BP produced could potentially display a new molecular recognition scaffold, and the phage titers thus provide an estimate of library diversity.
Testing transfer and display of His6 in Mtd position 349
Mtd 349 is located on the surface-exposed short alpha helix formed by G347 to S351 (Fig. 2A). A His6 epitope was inserted to test the tolerance of a short peptide in this position. The peptide transferred and successfully integrated into the vr by the DGR (Fig. 3C). DNA sequencing indicated that the transferred His6 peptide contained no mutations; however, 2 of the 12 variable codons in the surrounding endogenous scaffold were mutated in an adenine-error prone manner (n = 20).
Testing display of peptides in Mtd position 348
As described above, the tr encoding Mtd position 348 also tolerated heterologous inserts and was investigated for use in SMPL generation. Results include partial transfer of heterologous inserts at this position. For example, the 25-amino acids transferred in-frame into the expected location, but included only a sequence encoding the 19 of 25 C-terminal residues (Fig. 3D). Furthermore the adenines of the heterologous insert underwent the highly variable mutagenesis observed for the wild-type vr. Adenine replacement occurred more frequently at the 3′ terminus of the mtd, resulting in 59% of the endogenous adenines and 33% of the heterologous adenines mutated. The mutagenesis only altered the 3′ terminus of the insert, and did not alter mutable positions at the 5′ terminus of the insert. This experiment further demonstrates that a heterologous DNA sequence inserted into the tr can be accepted by the DGR for extensive mutagenesis during transfer to the correct position within the vr.
Testing insert size
To examine tolerance for a large DNA insert, the gene encoding the core of HIV-1 Nef (456 bp) was also inserted into positions 348 and 367 of the tr. Selections targeting a Nef ligand, p53, or anti-Nef antibody resulted in no increase in titers between wild-type phage and phage containing the Nef insert. Sequencing indicated that the Nef DNA had been excised from the tr. This result demonstrates the limits to the amount of DNA that can be maintained by the phage within the tr. Wild-type phage lacking the insert will replicate faster than phage with a large insert, and could quickly dominate the culture.
Model selections with the BP SMPL
Phage display selections tested the resilience of BP to the typical conditions used to identify binding ligands from protein libraries. TR and VR variants with His epitopes inserted were used to select for binding to immobilized metal affinity chromatography plates. After each round, the selected phages were propagated in Bvg− cells. Enrichment of the libraries of His6, His8 or His10-tagged phage was observed (Fig. 4A), and is being further studied. These results prove that selection conditions do not hinder the BP phage viability or infectivity. In addition, a phage-based ELISA examined display of a 2 × FLAG epitope at VR 367 (Table I and Fig. 4B). The observed binding to anti-FLAG antibodies demonstrates effective heterologous peptide display in this position.
To demonstrate selections with a protein target, tr mutants containing Asn4 or Asn8 inserts were used to target the HIV-1 protein gp41 (Table I). Selectants were propagated in Bvg+ bacteria and titered to determine the number of plaque forming units. Enrichments of >106-fold were observed from rounds 2 to 3, and a up to a 4-fold increase took place from round 3 to round 4. The wild type (BP) control showed no increase in titers (Fig. 4C, Table I); however, sequencing the selectants revealed loss of the insert and gain of mutations to the Mtd within the modified VR (n = 5). The experiments demonstrate the benefits of a nested combinatorial library to provide two potential solutions to molecular recognition challenges. If the library of inserts cannot find a solution, then the dynamically altered surrounding scaffold can offer potential binding affinity. To direct SMPL's toward mutagenesis of the inserted sequence, one could nucleate the diversity through incorporation of a known binding partner encoded in the tr, as demonstrated by experiments with the FLAG and His epitope tags (Fig. 4A and B).
Conclusion
The results reported here validate the approach of using a naturally occurring DGR to generate new protein libraries capable of binding a variety of targets. The caveats have also been documented here. For example, installation of the heterologous insert currently involves triparental mating using a suicide vector and helper plasmid. BioSafety Level 2 precautions are required for working with virulent (Bvg+) B.bronchseptica during the initial steps. Subsequent isolations and selections are done using virulence-repressed, non-pathogenic (Bvg−) B.bronchseptica (Liu et al., 2002; Doulatov et al., 2004). To date, the BP SMPL acts upon sequences encoding short peptides of up to ≈19 amino acids, and we have not observed the acceptance of sequences encoding larger proteins (e.g. Nef) by the mutagenesis system.
Effective molecular display systems require high levels of displayed protein diversity, a link from displayed molecule to its encoding information, and library members capable of binding to a range of different targets. Here, we demonstrate all such requirements in an SMPL system with the added capability for combinatorial self-synthesis. The successful identification of a loop region within the VR that can accept heterologous peptides sets the stage for display and engineering of peptides by SMPLs.
Supplementary data
Conflict of interest
None declared.
Funding
This work was supported by the National Institute of General Medical Sciences of the NIH (R01 GM078528-01) and a NIH pre-doctoral training grant to AML.
Supplementary Material
Acknowledgements
The authors thank Jeff F. Miller, Asher Hodes and Sergei Doulatov (UCLA) for supplying wild-type bacteria, Mtd antibodies and bacteriophage.
References
- Abagyan R., Batalov S., Cardozo T., Totrov M., Webber J., Zhou Y. Proteins. 1997:29–37. doi: 10.1002/(sici)1097-0134(1997)1+<29::aid-prot5>3.3.co;2-4. 1(Suppl) [DOI] [PubMed] [Google Scholar]
- Adams M.H. Bacteriophages. New York, NY: Interscience Publishers, Inc; 1959. [Google Scholar]
- Akerley B.J., Cotter P.A., Miller J.F. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- Akerley B.J., Miller J.F. Trends Microbiol. 1996;4:141–146. doi: 10.1016/0966-842x(96)10024-x. [DOI] [PubMed] [Google Scholar]
- Atwell S., Ultsch M., De Vos A.M., Wells J.A. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- Cheng J., Randall A.Z., Sweredoski M.J., Baldi P. Nucleic Acids Res. 2005;33:W72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.A., Miller J.F. In: Principles of Bacterial Pathogenesis. Groisman E.A, editor. San Diego: Academic Press; 2001. pp. 619–674. [Google Scholar]
- Dai W., Hodes A., Hui W.H., Gingery M., Miller J.F., Zhou Z.H. Proc. Natl Acad. Sci. U S A. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty P.S., Olsen M.J., Iverson B.L., Georgiou G. Protein Eng. 1999;12:613–621. doi: 10.1093/protein/12.7.613. [DOI] [PubMed] [Google Scholar]
- DeLano W.L., Lam J.W. Abstracts Pap. Am. Chem. Soc. 2005;230:U131–U1372. [Google Scholar]
- Diaz J.E., Howard B.E., Neubauer M.S., Olszewski A., Weiss G.A. Curr. Issues Mol. Biol. 2003;5:129–145. [PubMed] [Google Scholar]
- Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R.W., Zimmerly S., Miller J.F. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- Doyon J.B., Snyder T.M., Liu D.R. J. Am. Chem. Soc. 2003;125:12372–12373. doi: 10.1021/ja036065u. [DOI] [PubMed] [Google Scholar]
- Edwards R.A., Keller L.H., Schifferli D.M. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- Figurski D.H., Helinski D.R. Proc. Natl Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Tse L.V., Barbalat R., Sivaamnuaiphorn S., Xu M., Doulatov S., Miller J.F. Mol. Cell. 2008;31:813–823. doi: 10.1016/j.molcel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaja T.L., Narasimhamurthy S., Payan D.G., Anderson D.C. Chem. Biol. 2000;7:515–527. doi: 10.1016/s1074-5521(00)00137-x. [DOI] [PubMed] [Google Scholar]
- Hanes J., Plückthun A. Proc. Natl Acad. Sci. USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe J.W., Kay B.K. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Deora R., Doulatov S.R., et al. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- Liu M., Gingery M., Doulatov S.R., et al. J. Bacteriol. 2004;186:1503–1517. doi: 10.1128/JB.186.5.1503-1517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks K.M., Rosinov M., Nolan G.P. Chem. Biol. 2004;11:347–356. doi: 10.1016/j.chembiol.2004.03.004. [DOI] [PubMed] [Google Scholar]
- McMahon S.A., Miller J.L., Lawton J.A., et al. Nat. Struct. Mol. Biol. 2005;12:886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- Nemazee D. Nat. Rev. Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- Pedersen H., Holder S., Sutherlin D.P., Schwitter U., King D.S., Schultz P.G. Proc. Natl Acad. Sci. USA. 1998;95:10523–10528. doi: 10.1073/pnas.95.18.10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.K., Smith G.P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Sidhu S.S. Curr. Opin. Biotechnol. 2000;11:610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- Sidhu S.S., Weiss Gregory A. In: Phage Display. Clackson T., Lowman Henry B., editors. New York: Oxford University Press; 2004. pp. 27–41. [Google Scholar]
- Smothers J.F., Henikoff S., Carter P. Science. 2002;298:621–622. doi: 10.1126/science.298.5593.621. [DOI] [PubMed] [Google Scholar]
- Stibitz S. Gene. 1998;208:183–189. doi: 10.1016/s0378-1119(97)00643-4. [DOI] [PubMed] [Google Scholar]
- Uhl M.A., Miller J.F. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- Wilson D.S., Keefe A.D., Szostak J.W. Proc. Natl Acad. Sci. USA. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung Y.A., Wittrup K.D. Biotechnol. Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.