Abstract
SIRT1, a highly conserved NAD+-dependent protein deacetylase, is a key metabolic sensor that directly links nutrient signals to animal metabolic homeostasis. Although SIRT1 has been implicated in a number of hepatic metabolic processes, the mechanisms by which hepatic SIRT1 modulates bile acid metabolism are still not well understood. Here we report that deletion of hepatic SIRT1 reduces the expression of farnesoid X receptor (FXR), a nuclear receptor that regulates bile acid homeostasis. We provide evidence that SIRT1 regulates the expression of FXR through hepatocyte nuclear factor 1α (HNF1α). SIRT1 deficiency in hepatocytes leads to decreased binding of HNF1α to the FXR promoter. Furthermore, we show that hepatocyte-specific deletion of SIRT1 leads to derangements in bile acid metabolism, predisposing the mice to development of cholesterol gallstones on a lithogenic diet. Taken together, our findings indicate that SIRT1 plays a vital role in the regulation of hepatic bile acid homeostasis through the HNF1α/FXR signaling pathway.
INTRODUCTION
SIRT1 is a mammalian member of the silent information regulator 2 (Sir2) family of proteins, also known as sirtuins (7). First identified in yeast as key components in gene silencing complexes (18), sirtuins have been increasingly recognized as crucial regulators for a variety of cellular processes, ranging from energy metabolism and stress response to tumorigenesis and aging (6). The mammalian genome encodes seven sirtuins, SIRT1 to SIRT7 (15). As the most conserved mammalian sirtuin, SIRT1 couples the deacetylation of numerous transcription factors and cofactors, including p53, E2F1, NF-κB, FOXO, peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α), c-myc, hypoxia-inducible factor 1 (HIF-1), HIF-2α, heat shock factor 1 (HSF1), liver X receptor (LXR), farnesoid X receptor (FXR), CLOCK and PER2, and TORC2 (2, 9, 13, 21, 26, 28, 29, 32, 34, 35, 42, 49, 55, 58, 59), to the hydrolysis of NAD+. Therefore, SIRT1 has been considered as a metabolic sensor that directly links cellular metabolic status to gene expression regulation, playing an important role in a number of prosurvival and metabolic activities (19).
In the liver, the central metabolic organ that controls key aspects of nutrient metabolism (48), SIRT1 has been shown to regulate metabolism of both glucose and lipids (45). For instance, SIRT1 inhibits TORC2, a key mediator of early phase gluconeogenesis, leading to decreased gluconeogenesis during the short-term fasting phase (28). Prolonged fasting, on the other hand, increases SIRT1-mediated deacetylation and activation of PGC-1α, an essential coactivator for a number of transcription factors, resulting in increased fatty acid oxidation and improved glucose homeostasis (41, 42). Consistently, adenoviral knockdown of SIRT1 reduces expression of fatty acid β-oxidation genes in the liver of fasted mice (43). Specific deletion of the exon 4 of the hepatic mouse SIRT1 gene, which results in a truncated, nonfunctional SIRT1 protein, impairs peroxisome proliferator-activated receptor α (PPARα) activity and fatty acid β-oxidation, thereby increasing the susceptibility of mice to high-fat diet-induced hepatic steatosis and hepatic inflammation (41). Furthermore, a complete deletion of hepatic SIRT1 by floxing exons 5 and 6 leads to the development of liver steatosis, hyperglycemia, oxidative damage, and insulin resistance, even on a normal chow diet (53, 54). Conversely, hepatic overexpression of SIRT1 mediated by adenovirus attenuates hepatic steatosis and endoplasmic reticulum (ER) stress and restores glucose homeostasis in mice (27). In addition to glucose and fatty acid metabolism, SIRT1 has also been reported to regulate hepatic lipid homeostasis through a number of nuclear receptors and transcription factors (21, 26, 40, 51).
In this report, we show that hepatic SIRT1 modulates bile acid metabolism through regulation of farnesoid X receptor (FXR) expression. FXR is an important nuclear receptor in the regulation of systemic cholesterol and bile acid metabolism (12, 20). A recent report by Kemper et al. has shown that SIRT1 modulates the FXR signaling through direct deacetylation of this transcription factor in a mouse model in which hepatic SIRT1 was knocked-down by short hairpin RNA (shRNA) (21). Using a liver-specific SIRT1 knockout mouse model (SIRT1 LKO), we show here that permanent deletion of hepatic SIRT1 with the flox/albumin-Cre system decreases FXR signaling largely through reduced activity of hepatocyte nuclear factor 1α (HNF1α), a homeodomain-containing transcription factor that plays an important role in the transcriptional regulation of FXR (46). We found that deficiency of SIRT1 in the liver decreases the HNF1α recruitment to the FXR promoter and reduces the expression of FXR, resulting in impaired transport of biliary bile acids and phospholipids and increased incidence of cholesterol gallstones.
MATERIALS AND METHODS
Animal experiments.
Liver-specific SIRT1 knockout (SIRT1 LKO) mice in a C57BL/6 background were generated as described previously (41). Nine- to 10-month-old SIRT1 LKO mice and their age-matched littermate Lox controls (albumin-Cre negative, SIRT1flox/flox) were fed ad libitum either a standard laboratory chow diet or a lithogenic diet (D12383; Research Diets) for 6 weeks. All animal experiments were conducted in accordance with guidelines of the National Institute of Environmental Health Sciences (NIEHS)/NIH Animal Care and Use Committee.
Histological and biochemical analysis.
Paraffin-embedded liver sections were stained with hematoxylin and eosin for morphology. Serum low-density lipoprotein (LDL), high-density lipoprotein (HDL), and total cholesterol and triglycerides were measured using commercially available kits (Wako and Sigma). Serum insulin and leptin levels were measured by enzyme-linked immunosorbent assay (ELISA) (Meso scale discovery). Serum alanine transaminase (ALT) activities were measured using the ALT kit from Catechem.
To examine the biliary lipid profiles of control and SIRT1 LKO mice, bile was collected from the gallbladder, and then total bile acids were measured with the total bile acid kit based on 3α-hydroxysteroid dehydrogenase (Diazyme Laboratories). Biliary phospholipids and total cholesterol were determined using commercial kits from Wako. The cholesterol saturation indices (CSI) were then calculated from the critical tables in reference 10.
To determine the fecal bile acid outputs, feces were collected from individually housed mice over 24 h and bile acids from feces were extracted with 75% ethanol at 50°C for 2 h, followed by centrifugation at 1,500 × g for 10 min. Bile acids were then measured in the resulting supernatants.
Cell culture.
HEK293T cells stably infected by pSuper or pSuper-SIRT1 RNA interference (RNAi) were described previously (26). Mouse primary hepatocytes were isolated from control or SIRT1 LKO mice using collagenase perfusion, seeded on collagen-coated plates in seeding medium (high-glucose Dulbecco's modified Eagle's medium [DMEM], 10% fetal bovine serum [FBS], 100 nM insulin, 1 μM dexamethasone), and maintained in maintenance medium (high-glucose DMEM, 0.1% bovine serum albumin). To induce the expression of FXR target genes, primary hepatocytes were treated with dimethyl sulfoxide (DMSO) or 1 μM GW4064 in high-glucose medium for 24 h.
Western blot analysis, coimmunoprecipitation, and chromatin immunoprecipitation (ChIP) analysis.
Liver total-cell homogenates were prepared in SDS buffer (50 mM Tris-HCL [pH 6.8], 4% SDS), incubated at 100°C for 10 min, and then immunoblotted using antibodies against SIRT1 (Sigma), HNF1α (Santa Cruz Biotechnology), FXR (Santa Cruz Biotechnology), and actin.
For immunoprecipitation between SIRT1 and HA-HNF1α, HEK293T cells were transfected with the constructs indicated. Cells were then harvested 48 h later, cell lysates were prepared in NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% NP-40) containing Complete protease, and phosphatase inhibitors (Roche) were immunoprecipitated with antihemagglutinin (anti-HA) antibodies (Santa Cruz Biotechnology).
To determine the acetylation levels of FXR protein in liver, 1 mg of protein of liver extracts from control or SIRT1 LKO mice was incubated for 3 h with 1 μg of FXR antibody (goat polyclonal, sc-1204; Santa Cruz Biotechnology) under stringent conditions with SDS-containing radioimmunoprecipitation assay (RIPA) buffer. Acetylation levels of endogenous FXR in the immunoprecipitates were detected with anti-acetyl-Lys antibody (Cell Signaling). The membrane was stripped and reprobed with anti-FXR antibodies (mouse monoclonal, sc-25309; Santa Cruz Biotechnology).
Chromatin immunoprecipitation (ChIP) analysis was performed essentially as described by Upstate Biotechnology with modifications. Briefly, cells were cross-linked and harvested in IP buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and Complete protease inhibitor mixture). Chromatins were then sonicated to an average length of 200 to 500 bp and immunoprecipitated with antibodies against FXR, HNF1α (Santa Cruz biotechnology), SIRT1 (Millipore), or normal rabbit IgG. DNA fragments were subjected to quantitative PCR (qPCR) using primers flanking FXRE on the small heterodimer partner (SHP) promoter or different regions on the FXR promoter.
RNA analysis.
Total RNA was isolated from cells or tissues using TRIzol (Invitrogen) and the Qiagen RNeasy minikit (Qiagen). For real-time qPCR, cDNA was synthesized with the ABI reverse transcriptase kit and analyzed using SYBR green supermix (Applied Biosystems). All data were normalized to lamin A expression.
Luciferase assay.
For transactivation experiments, the mouse FXR promoter (fragment from −2644 to +149) was cloned into the pGL3 basic vector (Promega). Hepa1-6 cells were then transfected with FXR firefly luciferase reporter and control pRL-CMV (cytomegalovirus) reporter (Renilla luciferase; Promega), together with the indicated constructs, and were cultured for 24 h. Luciferase activity was measured using the dual-luciferase reporter assay system (Promega). The final firefly luciferase activity was normalized to the coexpressed Renilla luciferase activity.
Statistical analysis.
Values are expressed as means ± standard errors of means (SEM). Significant differences between the means were analyzed by two-tailed, unpaired Student's t test, and differences were considered significant at P < 0.05.
RESULTS
Hepatic deletion of SIRT1 reduces the expression of FXR.
To examine the function of SIRT1 in the liver, we previously generated liver-specific SIRT1 knockout (SIRT1 LKO) mice and analyzed the hepatic transcriptional expression profiles of these mice on a chow diet by microarray analyses (41). Interestingly, two related pathways that were significantly affected in the SIRT1 LKO mice were hepatic cholestasis and FXR/RXR activation (see Fig. S1 in the supplemental material). Among genes involved in these two pathways, their products Abcb4 (−1.967), Abcc3 (−1.734), Abcg8 (−1.153), and Slc10a2 (−2.010) are transporters that mediate the uptake and efflux of bile acids and cholesterol (see Tables S1 and S2 in the supplemental material), and Abcb4 is a direct transcriptional target of FXR.
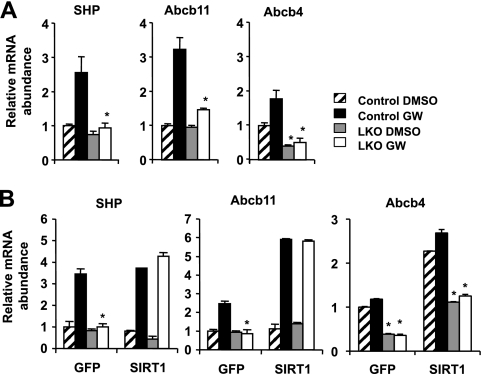
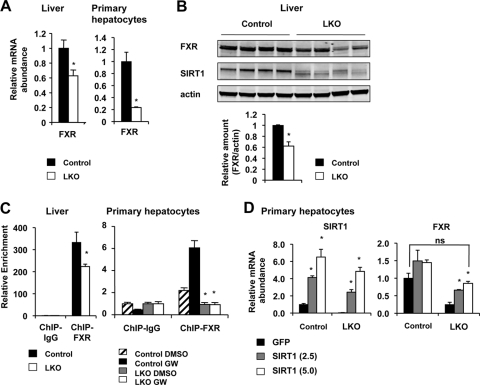
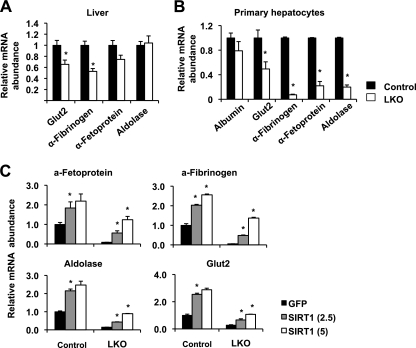
To confirm our microarray data that SIRT1 deficiency in the liver results in decreased FXR signaling, we isolated primary hepatocytes from Lox control or SIRT1 LKO mice and treated them with FXR agonist GW4064 (GW). As shown in Fig. 1A, the induction of a number of FXR target genes, including the small heterodimer partner (SHP), Abcb11, and Abcb4 genes, was significantly blunted in the SIRT1-deficient hepatocytes. Moreover, lentivirus-mediated overexpression of SIRT1 in LKO hepatocytes completely restored the mRNA levels of these genes (Fig. 1B). These data indicate that SIRT1 positively regulates FXR signaling directly in hepatocytes. To dissect the molecular mechanism underlying this regulation, we analyzed the expression levels of FXR in hepatocytes and livers. As shown in Fig. 2A and B, both the mRNA and protein levels of FXR were significantly decreased in SIRT1-deficient livers and primary hepatocytes. Furthermore, the chromatin-associated FXR levels at the FXRE of SHP were significantly decreased in the livers of SIRT1 LKO mice as well as in SIRT1-deficient primary hepatocytes (Fig. 2C). These observations suggest that SIRT1 may positively regulate the transcription of FXR and that the reduced FXR signaling in the SIRT1 LKO mice may be in part due to decreased expression of FXR. In support of our hypothesis, overexpression of SIRT1 in the SIRT1-deficient hepatocytes resulted in a dose-dependent increase of FXR mRNA levels and further stimulated the expression of FXR in the control hepatocytes (Fig. 2D).
Fig 1.
Loss of SIRT1 reduces the activity of FXR signaling pathway in primary hepatocytes. (A) SIRT1 deficiency in primary hepatocytes reduces the induction of FXR target genes by GW4064. *, P < 0.05. (B) Overexpression of SIRT1 in primary hepatocytes restores the expression of FXR targets. Primary hepatocytes from control and SIRT1 LKO mice were infected with lentiviruses expressing GFP or SIRT1. The mRNA levels of FXR target genes were analyzed by qPCR. *, P < 0.05.
Fig 2.
Loss of hepatic SIRT1 decreases the expression of FXR. (A) SIRT1 deficiency leads to reduced mRNA levels of FXR in the liver (n = 11) and primary hepatocytes (n = 3). *, P < 0.05. (B) SIRT1 deficiency results in reduced FXR protein levels in the liver (n = 4). *, P < 0.05. (C) Reduced recruitment of FXR to the FXRE on the promoter of SHP gene in the SIRT1-deficient livers and primary hepatocytes (n = 3). *, P < 0.05. (D) Overexpression of SIRT1 in SIRT1-deficient primary hepatocytes restores the expression of FXR. Primary hepatocytes from control and SIRT1 LKO mice were infected with lentiviruses expressing GFP or increased doses of SIRT1 (multiplicities of infection [MOI] = 2.5 and 5, respectively). The expression levels of SIRT1 and FXR were determined by qPCR. *, P < 0.05.
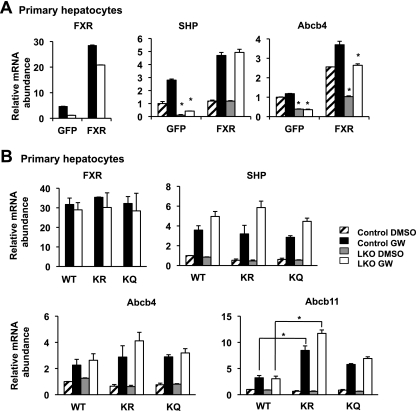
FXR has recently been reported as an acetylated transcription factor (21). It has been shown that acetylation of FXR by the acetyltransferase p300 leads to decreased heterodimerization with its partner, RXRα, resulting in reduced DNA binding and transactivation activity. SIRT1, on the other hand, is able to deacetylate FXR, thereby activating its activity (21). To test the importance of the observed transcriptional regulation of FXR by SIRT1 (Fig. 2) relative to the previously defined role of SIRT1 in enhancing the transactivation activity of FXR through deacetylation (21), we analyzed the transactivation activity of exogenous murine FXR protein in control and SIRT1-deficient primary hepatocytes. As shown in Fig. 3A, lentiviral expression of FXR in the SIRT1 KO hepatocytes completely rescued the deficient expression of SHP and almost completely recovered the levels of Abcb4, indicating that the transactivation activity of exogenous wild-type (WT) FXR protein is almost normal in the SIRT1-deficient hepatocytes. To further assess the transactivation activities of acetylated-FXR and deacetylated-FXR proteins in primary hepatocytes, we generated lentiviruses expressing mutant FXR proteins in which the previously identified lysine acetylation sites were mutated either to arginine (K168R/K228R [KR]) to mimic the deacetylation protein or to glutamine (K168Q/K228Q [KQ]) to mimic the acetylated FXR protein. The transactivation activities of these mutants were then analyzed in control and SIRT1 KO primary hepatocytes. As shown in Fig. 3B, the WT and mutant FXR proteins had comparable activities in both control and SIRT1-deficient hepatocytes on two of FXR target genes, the SHP and Abcb4 genes. However, the deacetylation-mimetic, KR mutant protein displayed significantly increased activity on Abcb11. These observations suggest that acetylation status of FXR impacts its transcriptional activity only on some target genes. Taken together, our data demonstrate that permanent deletion of SIRT1 in hepatocytes affects the activity of FXR largely through the transcriptional regulation of its expression.
Fig 3.
Loss of hepatic SIRT1 reduces the FXR signaling pathway primarily through transcriptional regulation. (A) Lentivirus-mediated expression of FXR in the SIRT1 KO hepatocytes rescues the deficient expression of FXR targets. Primary hepatocytes from control and SIRT1 LKO mice were infected with lentiviruses expressing GFP or FXR. The expression levels of FXR and FXR target genes were determined by qPCR. *, P < 0.05. (B) Transactivation activities of WT, KR, and KQ FXR proteins in control and SIRT1 LKO hepatocytes. *, P < 0.05. Primary hepatocytes from control and SIRT1 LKO mice were infected with lentiviruses expressing WT FXR, FXR KR, or FXR KQ mutant proteins. The expression levels of FXR and FXR target genes were determined by qPCR. *, P < 0.05.
SIRT1 regulates the expression of FXR through HNF1α.
As an important bile acid sensor that is critical for lipid and glucose metabolism, the expression of FXR is tightly controlled by an intricate regulatory network at multiple levels in response to various environmental stimuli. For example, the expression of FXR is under the control of HNF1α (46), a homeodomain-containing transcription factor that is essential for diverse metabolic processes in the pancreatic islets, liver, intestine, and kidney (25, 38, 39). The expression and transactivation activity of FXR are also regulated by an important coactivator for a number of transcription factors, peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α) (61), which is a direct deacetylation target of SIRT1 in the regulation of gluconeogenesis and fatty acid oxidation (41–43). FXR has also been reported to self-regulate its expression (reviewed by Eloranta and Kullak-Ublick [14]).
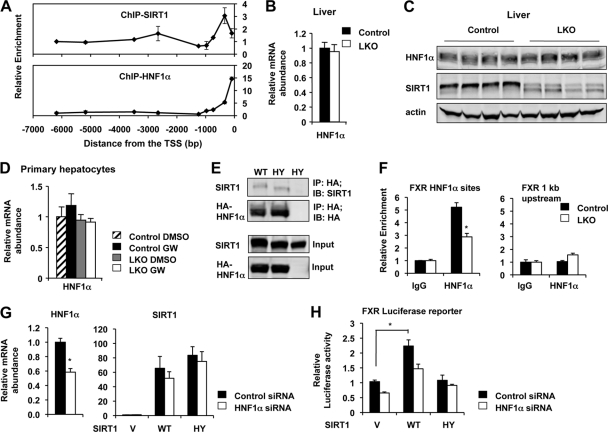
To dissect the molecular mechanisms by which loss of SIRT1 leads to the reduction of FXR expression, we analyzed the association of SIRT1 with the mouse FXR promoter. As shown in Fig. 4A, SIRT1 was relatively concentrated approximately 300 bp upstream of the transcription start site (TSS), where multiple binding sites of HNF1α were identified by the Genomatix MatInspector analyses (data not shown). Consistently, HNF1α was highly enriched near the TSS of the mouse FXR promoter (Fig. 4A). This observation suggests that SIRT1 may regulate the expression of FXR through modulation of HNF1α.
Fig 4.
SIRT1 regulates the expression of FXR through HNF1α. (A) SIRT1 is enriched on the HNF1α binding sites on the mouse FXR promoter. Primary hepatocytes from control and SIRT1 LKO mice were ChIPed with SIRT1 or HNF1α antibodies. DNA fragments were then subjected to qPCR using primers flanking the indicated regions on the FXR promoter. (B to D) SIRT1 deficiency does not cause defective expression of HNF1α in the liver (B and C) and primary hepatocytes (D). (E) SIRT1 interacts with HNF1α in HEK293T cells. Cell lysates from HEK293T cells expressing HA-HNF1α with wild-type (WT) or catalytically inactive (HY) SIRT1 were immunoprecipitated (IP) with anti-HA antibodies. (F) SIRT1 deficiency leads to decreased association of HNF1α with the HNF1α binding sites on the FXR promoter. Primary hepatocytes from control and SIRT1 LKO mice were ChIPed with IgG or HNF1α antibodies. (G and H) SIRT1 induces the expression of FXR through HNF1α in Hepa1-6 cells. Mouse hepatocyte Hepa1-6 cells were electroporated with negative control siRNA (control siRNA) or siRNA against HNF1α (HNF1α siRNA) and were then transfected with vector (V) or constructs expressing WT or HY SIRT1 together with mouse FXR promoter luciferase reporter. The expression levels of HNF1α and SIRT1 (G) were determined by qPCR, and the luciferase activity of FXR reporter was determined as described in Materials and Methods.
HNF1α is an important transcription factor that is involved in the differentiation program in several organs, including the liver, kidney, intestine, and pancreas. Haploinsufficiency of HNF1α in humans (56) and knockout of HNF1α in mice led to the development of diabetes, renal Fanconi syndrome, hepatic dysfunction, and hypercholesterolemia (25, 38, 39). Since both mRNA and protein levels of HNF1α were normal in SIRT1-deficient mice and primary hepatocytes (Fig. 4B to D), we speculated that SIRT1 might regulate the activity of this transcription factor at the posttranscriptional level, which then indirectly affects the expression of FXR. Consistent with this possibility, both WT and catalytically inactive (HY) SIRT1 were coimmunoprecipitated with HA-HNF1α in HEK293T cells (Fig. 4E). Moreover, the chromatin-associated HNF1α levels were significantly reduced in the SIRT1-deficient hepatocytes compared to the control hepatocytes in a chromatin immunoprecipitation assay (Fig. 4F), suggesting that deletion of SIRT1 in hepatocytes decreases the DNA binding affinity of HNF1α. To further confirm that SIRT1 regulates the expression of FXR through HNF1α, we electroporated negative control small interfering RNA (siRNA) or siRNA against HNF1α into the mouse hepatocyte Hepa1-6 cell line. We then transfected these siRNAs with vector (V) or constructs expressing WT or HY SIRT1 together with mouse FXR promoter luciferase reporter (Fig. 4G). As shown in Fig. 4H, in Hepa1-6 cells transfected with control siRNA, overexpression of WT SIRT1 but not the HY mutant significantly induced the luciferase reporter of FXR. However, this induction was decreased in HNF1α RNAi cells, suggesting that SIRT1 induces the expression of FXR in part through HNF1α.
In line with the observation that deletion of hepatic SIRT1 leads to reduced expression of HNF1α target gene FXR, SIRT1 deficiency in hepatocytes also reduced the expression of a number of other HNF1α target genes in both liver and primary hepatocytes (Fig. 5A and B). Furthermore, overexpression of SIRT1 in the KO hepatocytes led to a dose-dependent increase of the mRNA levels of HNF1α target genes and further stimulated the expression of these targets in the control hepatocytes (Fig. 5C). Collectively, these findings demonstrate that SIRT1 deficiency in hepatocytes impairs the expression of FXR through modulation of HNF1α transcriptional activity.
Fig 5.
Loss of SIRT1 in hepatocytes results in decreased expression of other HNF1α target genes. (A) SIRT1 deficiency in the liver leads to decreased expression of other HNF1α target genes (n = 11). *, P < 0.05. (B) SIRT1 deficiency leads to decreased expression of other HNF1α target genes in primary hepatocytes (n = 3) *, P < 0.05. (C) Overexpression of SIRT1 in SIRT1-deficient primary hepatocytes restores the expression of other HNF1α target genes in primary hepatocytes. *, P < 0.05. MOI = 2.5 and 5, respectively. a-Fetoprotein, α-fetoprotein; a-Fibrinogen, α-fibrinogen.
Hepatic deletion of SIRT1 leads to impaired hepatic lipid metabolism on the lithogenic diet.
Decreased activity of HNF1α in humans and mice has been associated with development of diabetes, hepatic dysfunction, and hypercholesterolemia (25, 38, 39, 56). Disruption of FXR in mice is also associated with the development of metabolic diseases, including diabetes and hypercholesterolemia (47). The decreased activity of HNF1α and reduced expression of FXR in the liver of the SIRT1 LKO mice suggest that these animals may display impaired metabolic functions mediated by these two factors.
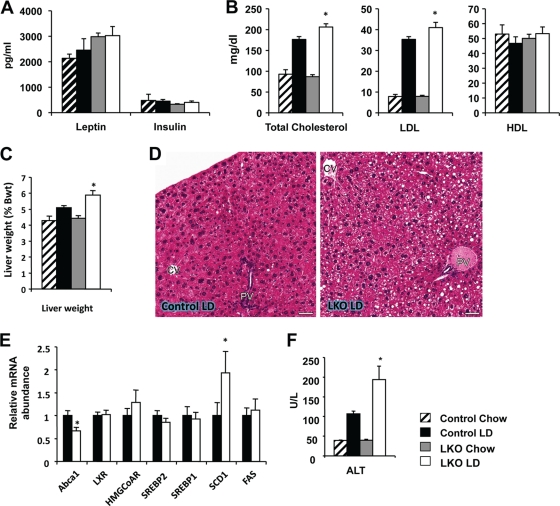
To examine the pathophysiological effects of blunted HNF1α and FXR signaling in vivo, we challenged the SIRT1 LKO and their Lox control littermates with a lithogenic diet that was high fat and high cholesterol and contained an additional 0.5% cholic acid. We have previously shown that deletion of SIRT1 in the liver results in decreased fatty acid oxidation, leading to a mild body weight gain and the development of hepatic steatosis and inflammation on a Western-style high-fat diet (41). When challenged with the lithogenic diet, SIRT1 LKO mice showed no obvious signs of body weight abnormality (data not shown) and had normal levels of serum insulin and leptin (Fig. 6A). However, they displayed mild but significant hypercholesterolemia, primarily through an increase of the LDL fraction in serum (Fig. 6B). They also displayed modest but significant hepatomegaly and had greater lipid accumulation in the liver (Fig. 6C and D). In line with these observations, SIRT1 LKO mice showed significantly altered expression of a couple of genes involved in cholesterol efflux and triglyceride biogenesis (Fig. 6E). Additionally, 6 weeks of lithogenic diet feeding also resulted in significantly greater liver damage in the SIRT1 LKO mice, as revealed by elevated serum alanine transaminase (ALT) activities (Fig. 6F).
Fig 6.
Hepatic loss of SIRT1 function impairs lipid homeostasis upon lithogenic diet feeding. After 6 weeks of lithogenic diet feeding, SIRT1LKO mice displayed normal serum hormonal levels (A) and a mild increase of serum total cholesterol and LDL levels (B) (n = 11). *, P < 0.05. (C and D) SIRT1 LKO mice display modest hepatomegaly (C) and hepatic steatosis (D) (n = 11). *, P < 0.05. Bars, 50 μm. (E) SIRT1 LKO mice display decreased expression of Abca1 and increased levels of SCD1 (n = 11). *, P < 0.05. (F) SIRT1 deficiency in the liver increases liver damage (n = 11). *, P < 0.05.
Hepatic deletion of SIRT1 impairs bile acid metabolism and induces formation of cholesterol gallstones on the lithogenic diet.
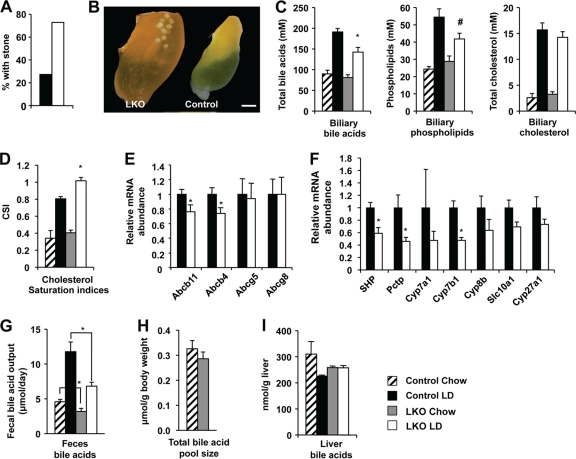
Disruption of FXR in mice has also been associated with the development of cholesterol gallstone disease upon lithogenic diet feeding (31). Strikingly, 6 weeks of lithogenic diet feeding induced formation of gallstones, which were visible at the macroscopic level (Fig. 7B) in 73% of SIRT1 LKO mice (Fig. 7A). In contrast, only 27% of the control mice acquired gallstones under the same conditions (Fig. 7A).
Fig 7.
Hepatic deletion of SIRT1 induces formation of cholesterol gallstones and impaired bile acid metabolism in mice upon a lithogenic diet feeding. (A) SIRT1 LKO mice display increased incidence of cholesterol gallstones (n = 11). The 9- to 10-month-old control and SIRT1 LKO mice were fed a lithogenic diet for 6 weeks. (B) Macroscopic appearance of gallbladders from control and SIRT1 LKO mice fed with the lithogenic diet for 6 weeks. Bar, 1 mm. (C) SIRT1 LKO mice display decreased biliary concentrations of bile acids and phospholipids (n = 8). *, P < 0.05; #, P = 0.068. (D) SIRT1 LKO mice have increased cholesterol saturation indices (CSI) in gallbladder bile (n = 8). *, P < 0.05. (E) SIRT1 deficiency reduces expression of bile acid and phospholipid transporters at the hepatocyte canalicular membrane (n = 11). *, P < 0.05. (F) Decreased expression of bile acid synthesis genes in the SIRT1 LKO mice (n = 11). *, P < 0.05. (G to I) SIRT1 LKO mice show decreased fecal bile acid output (G) but normal total bile acid pool size (H) and hepatic bile acids (I) (n = 5 to 6) *, P < 0.05.
Since the formation of gallstones is primarily determined by the relative biliary concentrations of bile salts/phospholipids and cholesterol (10, 11), which are under the control of FXR and LXR, respectively (31), we analyzed the biliary lipid profile in control and SIRT1 LKO mice. As shown in Fig. 7C, SIRT1 LKO mice showed reduced levels of biliary bile acids and phospholipids compared to control mice, whereas their biliary cholesterol concentrations were normal. As a result, their biliary cholesterol was significantly more saturated (Fig. 7D). These data suggest that the SIRT1 LKO mice have deficient FXR signaling on the lithogenic diet. In support of these observations, the expression levels of two FXR targets located at the outer leaflet of the hepatocyte canalicular membrane that are responsible for the biliary transport of bile salts and phospholipids (31), Abcb11 and Abcb4, were significantly lower in SIRT1 LKO mice compared to control mice (Fig. 7E). In contrast, the levels of two cholesterol transporters that are under the control of LXR, Abcg5 and Abcg8, were not changed (Fig. 7E). Together, these findings confirm that hepatic deletion of SIRT1 leads to defective FXR signaling, resulting in reduced biliary bile salt and phospholipid concentrations and increasing the risk of cholesterol gallstones while on the lithogenic diet.
In addition to regulating bile salts and phospholipid transport, HNF1α/FXR plays an important role in the regulation of bile acid synthesis, primarily through SHP, an odd member of the nuclear receptor superfamily (16, 20). Upon activation by bile acids, FXR induces the expression of SHP, which in turn binds to LXR, liver receptor homolog 1 (LRH-1), and possibly other nuclear receptors to attenuate further bile acid synthesis (44). Consistently, deletion of FXR in mice decreases the expression of SHP while increasing the mRNA levels of bile acid synthesis genes, such as Cyp7a1, Cyp8b1, and Cyp27a1 (31). Loss of SHP, on the other hand, partially impairs negative feedback regulation of bile acid synthesis (22, 52). The reduced levels of FXR and SHP suggest that SIRT1 LKO mice may also suffer from an abnormally high rate of bile acid synthesis. However, unexpectedly, the expression of a number of bile acid synthesis genes was lower in these mice (Fig. 7F). In line with this observation, their bile acid synthesis rates, based on the steady-state fecal bile acid output rate, were decreased by 30% and 43%, respectively, under both basal and lithogenic conditions (Fig. 7G). However, there were no significant alterations in the total bile acid pool size levels (Fig. 7H) and liver bile acid concentrations (Fig. 7H). These observations suggest that the FXR-SHP-bile acid synthesis feedback loop was impaired in the SIRT1 LKO mice.
DISCUSSION
As the best-studied member of the sirtuin family of antiaging proteins, SIRT1 is rising as an important therapeutic target for a number of age-associated diseases. While it has been reported that SIRT1 is a vital regulator in many aspects of hepatic lipid and glucose metabolism in response to different nutrient signals (19, 45) and that SIRT1 regulates the FXR signaling by direct deacetylation of this transcription factor (21), we show in the present study that hepatic SIRT1 modulates the message RNA levels of FXR through HNF1α. As a result, deletion of SIRT1 in the liver results in decreased HNF1α recruitment to the FXR promoter and reduced expression of FXR, leading to decreased transport of biliary bile acids and phospholipids and increased incidence of cholesterol gallstones. These observations uncover a previously unknown link between SIRT1, HNF1α, and transcriptional regulation of FXR expression, suggesting that hepatic SIRT1 may also be an important therapeutic target for cholesterol gallstone disease.
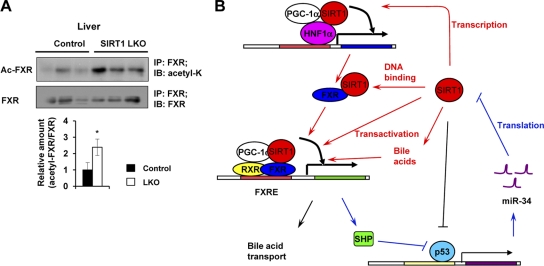
Several lines of evidence suggest that hepatic SIRT1 modulates the FXR signaling pathway predominantly at the transcriptional level in the SIRT1 LKO mouse model. For instance, the expression of FXR is decreased not only in the SIRT1-deficient liver, but also in the SIRT1-deficient hepatocytes (Fig. 2A), indicating that SIRT1 directly regulates FXR expression in a cell autonomous fashion. Moreover, overexpression of SIRT1 in primary hepatocytes induces the expression of FXR (Fig. 2D). More importantly, it appears that the transactivation activity of exogenous FXR is normal in the SIRT1-deficient hepatocytes, as lentiviral expression of FXR almost completely rescues the defective FXR activity in these cells (Fig. 3A). On the other hand, our results also point toward the involvement of additional mechanisms. As shown in Fig. 3A, putting back FXR did not completely restore the expression of all tested FXR downstream targets in the SIRT1-deficient hepatocytes. It appears that the acetylation status of the FXR proteins indeed affects their transactivation activities on some of the FXR targets, particularly in SIRT1-deficient hepatocytes (Fig. 3B). Furthermore, the FXR proteins are markedly hyperacetylated in the SIRT1 LKO mice (Fig. 8A). These observations indicate that SIRT1 also plays a role in the posttranslational activation of FXR through direct deacetylation of the receptor, thereby improving its DNA binding ability (21). In addition to HNF1α, the expression and transactivation activity of FXR are also regulated by a PGC-1α (61). We have previously shown that hepatocyte-specific deletion of SIRT1 leads to decreased coactivation activity of PGC-1α (41). Although our preliminary data indicate that PGC-1α is still capable of promoting the expression of FXR in the SIRT1-deficient hepatocytes (data not shown), the decreased activity of PGC-1α may partially contribute to the reduction of the FXR signaling in these cells. Interestingly, recent studies have demonstrated that the FXR-SHP pathway also positively regulates the translation of SIRT1 protein via the p53/miR-34a pathway (24, 57). It has been shown that SHP, one of the direct FXR targets, inhibits transactivation of transcription factor p53 and the expression of its downstream target, miR-34a, which in turn binds to the 3′ untranscribed region (3′ UTR) of SIRT1 mRNA, inhibiting the translation of SIRT1 protein (24, 57). Therefore, SIRT1 and the FXR signaling pathway mutually interact at multiple levels, coordinately regulating hepatic bile acid and cholesterol homeostasis (Fig. 8B).
Fig 8.
SIRT1 regulates FXR signaling at multiple levels. (A) Acetyl-FXR levels are significantly elevated in SIRT1 LKO mice. Total liver extracts from control and SIRT1 LKO mice were immunoprecipitated with goat anti-FXR antibodies and then immunoblotted with rabbit anti-acetyl-K antibodies or mouse anti-FXR antibodies (n = 3). *, P < 0.05. (B) The interaction network between SIRT1 and the FXR signaling pathway. SIRT1 modulates the activity of FXR signaling at multiple levels (red lines). First, SIRT1 regulates the expression of FXR through interaction with HNF1α, which may also involve coactivator PGC-1α. Second, SIRT1 deacetylates FXR, increasing its DNA binding affinity. Third, SIRT1 deacetylates PGC-1α, thereby activating the transactivation activity of FXR. Finally, SIRT1 stimulates the biosynthesis of bile acids, which in turn bind to FXR and induce its transcriptional activity. FXR, on the other hand, increases the translation of SIRT1 protein via SHP-mediated inhibition of p53 transactivation and thereby miR34a expression (blue lines). This positive feedback network plays an essential role in the regulation of bile acid metabolism. Alterations of these loops in the SIRT1 LKO mice lead to impairments in bile acid synthesis and transport, resulting in development of cholesterol gallstones.
Given the facts that HNF1α is paramount to normal functions of liver and pancreas and that human HNF1α is commonly mutated in patients with maturity onset diabetes of the young (MODY), which is characterized by severe insulin secretory defects (1, 56), the positive forward link between SIRT1, HNF1α, and FXR revealed in the present study is not altogether surprising. For example, it has been shown that increased dosage of SIRT1 in pancreatic β cells improves glucose tolerance and enhances insulin secretion in response to glucose (33), whereas deletion of SIRT1 impairs glucose-stimulated insulin secretion (8). Resveratrol, a polyphenol activator of SIRT1, potentiates glucose-stimulated insulin secretion in β cells (50). In addition, activation of SIRT1 by its activators in animals protects against high-fat-induced obesity and insulin resistance (4, 23, 30), and modest overexpression of SIRT1 resulted in a protective effect against high-fat-induced hepatic steatosis and glucose intolerance (3, 37). Our data suggest that activation of the HNF1α signaling pathway may partially underlie the antidiabetes action of SIRT1. Further exploration of this possibility may provide novel insights into SIRT1's function in whole-body glucose homeostasis. Our data, however, are in contrast to those reported in a recent study (17). Grimm et al. have shown that SIRT1 and HNF1α form a nutrient-sensitive complex, and this interaction suppresses the transcriptional activity of HNF1α on its target genes, particularly C-reactive protein (CRP), in hepatocytes. In addition, their data show that SIRT1 inhibits HNF1α activity on the CRP promoter through deacetylation of H4K16 instead of HNF1α itself (17). One possible factor contributing to the discrepancy between our observations and those of Grimm et al. may be the difference in environmental challenges in two studies. SIRT1 appears to suppress HNF1α and the production of CRP only under conditions of nutrient restriction (17), whereas SIRT1 activates the HNF1α/FXR pathway in hepatocytes regardless of nutrient status. In addition, the HNF1α-mediated gene expression involves formation of multiunit transcriptional complexes with other transcription factors. For instance, cytokine-driven expression of the CRP gene requires formation of c-Fos, STAT3, and the HNF1α transcriptional complex (36), among which c-Fos can be deacetylated and inhibited by SIRT1 (60). Therefore, it is possible that the net effect of SIRT1 on the expression of different HNF1α target genes is determined by the combination of different transcriptional partners.
How SIRT1 regulates the activity of HNF1α is still an ongoing study. Since the chromatin-associated HNF1α levels were significantly reduced in the livers of SIRT1 LKO mice (Fig. 4F), loss of SIRT1 may directly or indirectly decrease the DNA binding affinity of HNF1α. Additional experiments are needed to dissect the molecular mechanisms underlying this important modulation in vitro and in vivo.
The impaired FXR-SHP-bile acid synthesis feedback loop in the SIRT1 LKO mice suggests that hepatic deletion of SIRT1 may disrupt bile acid synthesis through FXR-independent mechanisms. For instance, increased liver damage in the SIRT1 LKO mice under the lithogenic diet may activate c-Jun N-terminal kinase (JNK), thereby inhibiting bile acid synthesis (22, 44, 52). Deletion of SIRT1 in the hepatocytes may also result in hepatic insulin resistance, a condition known to decrease the expression of the first committed enzyme in the acidic pathway of bile acid synthesis, Cyp7b1 (5). However, comparison of levels of phospho-JNK, a marker of the activated JNK pathway, in the liver extracts of control and SIRT1 LKO mice failed to reveal significant alterations of this signaling pathway (data not shown). Ser473 phosphorylation of Akt, a key molecule in the insulin signaling pathway, was also normal in the livers of SIRT1 LKO mice (data not shown). Therefore, additional studies are required to dissect the mechanism underlying this intriguing phenotype.
In summary, we have shown that hepatic SIRT1 plays an important role in the regulation of hepatic bile acid metabolism. Hepatic deletion of SIRT1 leads to an increased susceptibility to cholesterol gallstone disease through decreased HNF1α/FXR signaling. Our findings provide a direct link between SIRT1 and cholesterol gallstone disease and suggest that new therapeutic strategies designed to modulate SIRT1 activity may be beneficial for preventing formation of cholesterol gallstones as well as for other metabolic diseases associated with type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anton Jetten, Stavros Garantziotis, Darryl Zeldin, and members of the Li laboratory for critical reading of the manuscript. We thank Frederic Alt at Harvard Medical School for providing the SIRT1 exon 4 floxed allele. We also thank the NIEHS Laboratory of Experimental Pathology for histological staining and serum hormone ELISA and the NIEHS viral core facility for lentiviruses.
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, to X.L. (Z01 ES102205).
Footnotes
Published ahead of print 30 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Armendariz AD, Krauss RM. 2009. Hepatic nuclear factor 1-alpha: inflammation, genetics, and atherosclerosis. Curr. Opin. Lipidol. 20:106–111 [DOI] [PubMed] [Google Scholar]
- 2. Asher G, et al. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317–328 [DOI] [PubMed] [Google Scholar]
- 3. Banks AS, et al. 2008. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baur JA, et al. 2006. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biddinger SB, et al. 2008. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat. Med. 14:778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop NA, Guarente L. 2007. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 8:835–844 [DOI] [PubMed] [Google Scholar]
- 7. Blander G, Guarente L. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417–435 [DOI] [PubMed] [Google Scholar]
- 8. Bordone L, et al. 2006. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunet A, et al. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015 [DOI] [PubMed] [Google Scholar]
- 10. Carey MC. 1978. Critical tables for calculating the cholesterol saturation of native bile. J. Lipid Res. 19:945–955 [PubMed] [Google Scholar]
- 11. Carey MC, Small DM. 1978. The physical chemistry of cholesterol solubility in bile. Relationship to gallstone formation and dissolution in man. J. Clin. Invest. 61:998–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cariou B, Staels B. 2007. FXR: a promising target for the metabolic syndrome? Trends Pharmacol. Sci. 28:236–243 [DOI] [PubMed] [Google Scholar]
- 13. Dioum EM, et al. 2009. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 324:1289–1293 [DOI] [PubMed] [Google Scholar]
- 14. Eloranta JJ, Kullak-Ublick GA. 2005. Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch. Biochem. Biophys 433:397–412 [DOI] [PubMed] [Google Scholar]
- 15. Frye RA. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793–798 [DOI] [PubMed] [Google Scholar]
- 16. Goodwin B, et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517–526 [DOI] [PubMed] [Google Scholar]
- 17. Grimm AA, Brace CS, Wang T, Stormo GD, Imai SI. 2010. A nutrient-sensitive interaction between Sirt1 and HNF-1alpha regulates Crp expression. Aging Cell 10:305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021–1026 [PubMed] [Google Scholar]
- 19. Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5:253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalaany NY, Mangelsdorf DJ. 2006. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68:159–191 [DOI] [PubMed] [Google Scholar]
- 21. Kemper JK, et al. 2009. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 10:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kerr TA, et al. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2:713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lagouge M, et al. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- 24. Lee J, et al. 2010. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 285:12604–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee YH, Sauer B, Gonzalez FJ. 1998. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1alpha knockout mouse. Mol. Cell. Biol. 18:3059–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X, et al. 2007. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28:91–106 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, et al. 2011. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 25:1664–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, et al. 2008. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo J, et al. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148 [DOI] [PubMed] [Google Scholar]
- 30. Milne JC, et al. 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moschetta A, Bookout AL, Mangelsdorf DJ. 2004. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat. Med. 10:1352–1358 [DOI] [PubMed] [Google Scholar]
- 32. Motta MC, et al. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551–563 [DOI] [PubMed] [Google Scholar]
- 33. Moynihan KA, et al. 2005. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2:105–117 [DOI] [PubMed] [Google Scholar]
- 34. Nahle Z, et al. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859–864 [DOI] [PubMed] [Google Scholar]
- 35. Nakahata Y, et al. 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishikawa T, et al. 2008. Transcriptional complex formation of c-Fos, STAT3, and hepatocyte NF-1 alpha is essential for cytokine-driven C-reactive protein gene expression. J. Immunol. 180:3492–3501 [DOI] [PubMed] [Google Scholar]
- 37. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. 2008. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U. S. A. 105:9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pontoglio M, et al. 1996. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84:575–585 [DOI] [PubMed] [Google Scholar]
- 39. Pontoglio M, et al. 1998. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J. Clin. Invest. 101:2215–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ponugoti B, et al. 2010. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 285:33959–33970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Purushotham A, et al. 2009. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9:327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodgers JT, et al. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- 43. Rodgers JT, Puigserver P. 2007. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U. S. A. 104:12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schoonjans K, Auwerx J. 2002. A sharper image of SHP. Nat. Med. 8:789–791 [DOI] [PubMed] [Google Scholar]
- 45. Schug TT, Li X. 2011. Sirtuin 1 in lipid metabolism and obesity. Ann. Med. 43:198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shih DQ, et al. 2001. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27:375–382 [DOI] [PubMed] [Google Scholar]
- 47. Sinal CJ, et al. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- 48. van den Berghe G. 1991. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J. Inherit. Metab. Dis. 14:407–420 [DOI] [PubMed] [Google Scholar]
- 49. Vaziri H, et al. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- 50. Vetterli L, Brun T, Giovannoni L, Bosco D, Maechler P. 2010. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E beta-cells and human islets through Sirt1 dependent mechanism. J. Biol. Chem. 286:6049–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker AK, et al. 2010. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24:1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L, et al. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2:721–731 [DOI] [PubMed] [Google Scholar]
- 53. Wang RH, et al. 2011. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 121:4477–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang RH, Li C, Deng CX. 2010. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int. J. Biol. Sci. 6:682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323:1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamagata K, et al. 1996. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384:455–458 [DOI] [PubMed] [Google Scholar]
- 57. Yamakuchi M, Ferlito M, Lowenstein CJ. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 105:13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yeung F, et al. 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan J, Minter-Dykhouse K, Lou Z. 2009. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 185:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang R, et al. 2010. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J. Biol. Chem. 285:7097–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. 2004. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 18:157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.