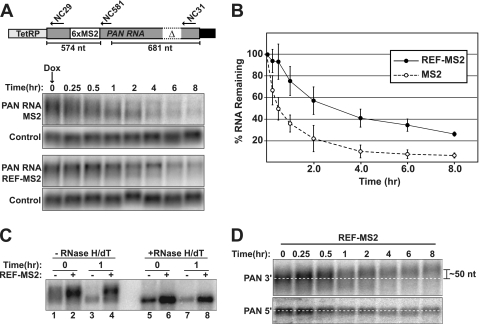
Fig 6.
REF/Aly promotes PAN RNA stability. (A) (Top) Schematic diagram of the TetRP-PANΔENE-6xMS2 construct. Positions of oligonucleotides (arrows) for RNase H-targeted cleavage (NC581) as well as probes for detection of the 5′ (NC29) and 3′ (NC31) ends of PAN RNA are shown. The schematic is not to scale. (Bottom) Northern blots from an in vivo RNA decay experiment. 293TOA cells were cotransfected with TetRP-PANΔENE-6MS2 and MS2 (upper panels) or REF-MS2 (lower panels). After transcription shutoff by addition of Dox to the medium, RNA was collected at the indicated intervals. Blots were probed first for PAN RNA and then for an endogenous loading control (7SK RNA). (B) Decay curve of PANΔENE with or without REF-MS2. PAN RNA signals were first normalized to the loading control and expressed as a percentage of the time-zero samples. Each data point is the average of three experiments, with the error bars representing the standard deviation. (C) Northern blot of 0- and 1-h samples from an RNA decay experiment with either MS2 alone (−) or REF-MS2 (+). The same samples were treated with RNase H and oligo(dT) to remove the poly(A) tails and run on the same gel. (D) REF-MS2 promotes PAN RNA hyperadenylation subsequent to transcription shutoff. Total RNA from a REF-MS2 transcription shutoff experiment was cleaved with RNase H and NC581 and subjected to Northern blotting. (Top) The 3′ end of the cleaved PAN RNA was detected with probe NC31. (Bottom) The 5′ PAN RNA fragment was detected on the same blot using probe NC29. The dashed white lines serve as guides to show that the mean length of the 3′ end increases while the 5′ end remains constant. The mean poly(A) tail lengths were estimated to be ∼150 nt at 0 h and ∼200 nt at 8 h (data not shown). The positions of the oligonucleotides used for RNase H cleavage and RNA detection are depicted in panel A.