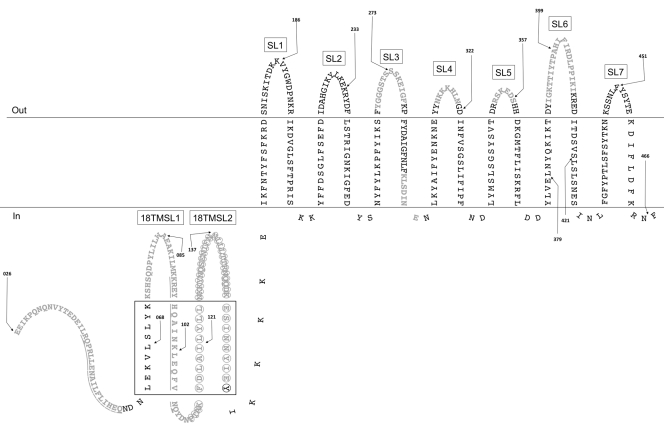
Fig 1.
NilB topology predicted by the PROFtmb (9) hidden Markov model. The model presented is similar to the model predicted by B2TMR-HMM (see Fig. S2 in the supplemental material and reference 36). While the PROFtmb model predicts that the ∼130 N-terminal amino acids are periplasmic, TMBB-PRED predicts that ∼100 of these amino acids form four additional TM domains (5, 6) (drawn in the periplasm in the same manner as TM domains and surface loops; the black box denotes this region). Only mature NilB is pictured (as predicted by SignalP 3.0 [8]), and numbering is according to that of immature NilB. Black arrows, FLAG tag insertions (before indicated amino acid); gray text, deletions (underlining distinguishes adjacent deletions); circles, the predicted TPR domain; SL, surface loop; 18TMSL, surface loop unique to the 18-TM strand model.