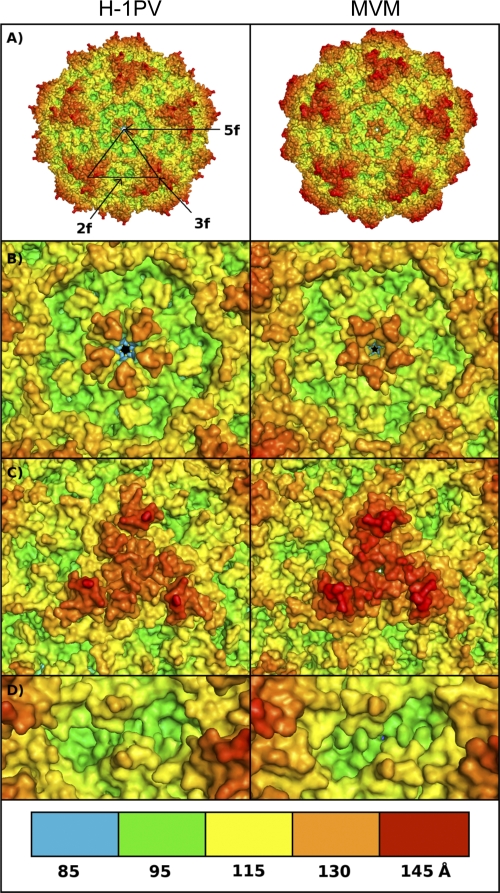
Fig 1.
In silico model of H-1PV and MVM parvovirus capsids. (A) The capsid of H-1PV (left) was modeled using the available crystal structure model of the MVM (right) capsid as a template. The approximate positions of icosahedral 5-fold (5f), 3-fold (3f), and 2-fold (2f) axes of symmetry are shown for a viral asymmetric unit. (B to D) Close-up views of the H-1PV (left) and MVM (right) capsid surfaces at the 5-fold (B), 3-fold (C), and 2-fold (D) icosahedral axes. The panel at the bottom depicts the color range (in Å) for the depth-cued distances from the viral center of the particles. The color coding of the particles was generated by an in-house-developed algorithm, and images were generated with PyMol software as indicated in Materials and Methods.