Fig 2.
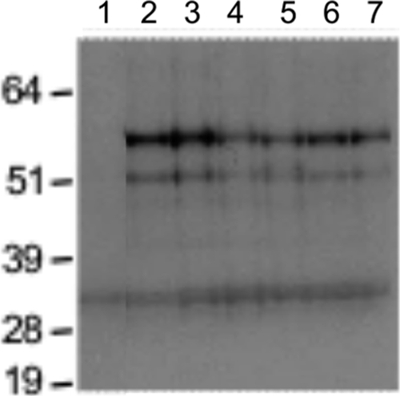
Protein truncation test for tirant env in D. simulans natural strains. Western blotting of the products from in vitro transcription and translation. Lanes: 1, D. pseudoobscura negative control; 2, D. melanogaster strain from Senegal; 3 to 7, D. simulans strains from Mayotte, Brazzaville, Zimbabwe, Makindu, and Chicharo, respectively. The molecular mass ladder (in kDa) is included on the left side of the image. All of the tested D. simulans natural populations displayed identical profiles that corresponded to a potentially functional env ORF. The fainter, lower 33-kDa band is not tirant specific because we also found this band in the D. pseudoobscura negative control (a species devoid of tirant insertions).
