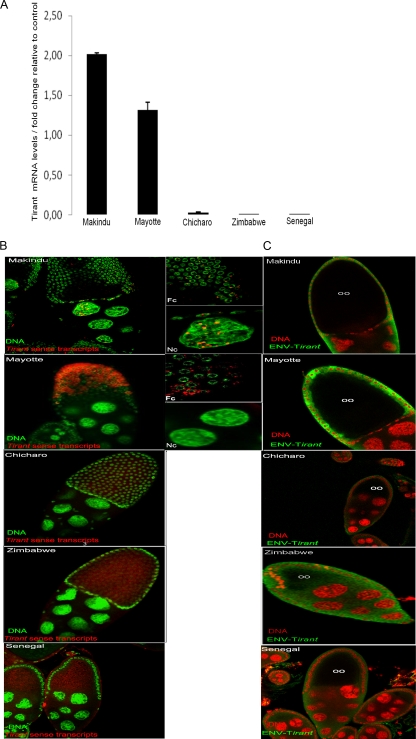
Fig 4.
Expression of the tirant env gene in the ovaries of natural strains of D. simulans. (A) RT-qPCR of the ovaries. The transcript levels were estimated relative to those of the rp49 gene. Significant levels of tirant env transcripts were detected in the Makindu and Mayotte populations. D. melanogaster Senegal was used as a control strain. (B) In situ hybridization of tirant transcripts (red). DNA is labeled in green (Sytox Green). Fc, follicle cells; Nc, nurse cells. (Left) View of ovary chambers (magnification, ×40); (right) details of the left panel. tirant transcripts were found in the nurse cells and follicle cells of the Makindu strain and in the follicle cells of the Mayotte strain. No staining was observed in the other strains. (C) Detection of tirant Env protein by immunochemistry (green) in the Mayotte strain. DNA is stained in red (propidium iodide). oo, oocyte. The green halos in the other strains do not represent Env staining but rather correspond to autofluorescence, as they were also observed following staining with preimmune serum.