Abstract
Expression arrays were used to identify 4 putative oxidoreductases that were upregulated (>3-fold) by furfural (15 mM, 15 min). Plasmid expression of one (ucpA) increased furan tolerance in ethanologenic strain LY180 and wild-type strain W. Deleting ucpA decreased furfural tolerance. Although the mechanism remains unknown, the cryptic ucpA gene is now associated with a phenotype: furan resistance.
TEXT
Furfural is an inhibitory side product formed by the dehydration of pentose sugars during dilute acid pretreatment of lignocellulosic biomass (1, 3, 14, 20). An analogous compound, 5-hydroxymethylfurfural (5-HMF), is produced from hexose sugars such as fructose. These furans inhibit the growth and fermentation of biocatalysts such as yeasts (2, 9–13) and ethanologenic Escherichia coli (7, 14, 15, 28, 29), complicating fermentation processes.
Furan addition to E. coli fermentations results in an initial period of slow growth or lag, during which furans are reduced to alcohols and remain in the broth (11). Furfural-resistant mutants of ethanologenic E. coli LY180 have been isolated and characterized (17, 18, 26). Partial resistance to low concentrations of furfural resulted from the silencing of yqhD, a furfural-induced NADPH-dependent furfural oxidoreductase (17, 18, 26). Although multiple NADPH-dependent furfural reductases are present in E. coli and conversion of furfural to the less toxic alcohol is generally regarded as beneficial, the unusually low Km of YqhD for NADPH (9 μM) is proposed to inhibit growth by depleting NADPH (17, 18). Furfural tolerance was improved by expression of fucO (propanediol oxidoreductase), an NADH-dependent furfural reductase (27) that normally functions during fucose catabolism (4). The use of NADH as the electron donor is of particular interest, because NADH is more abundant during fermentation (6, 27) and because its use for furfural reduction would not compete with biosynthesis.
The expression of all known furfural reductases in E. coli (YqhD, DkgA, and FucO) is upregulated by furfural (17). To identify additional NADH-dependent furfural reductases, mRNA expression levels in control cells (LY180) were compared to cells exposed to furfural (15 mM) for 15 min as previously described (17). Of the 261 genes with a 3-fold or higher expression, four were oxidoreductases (aldA, xdhABC, yeiTA, and ucpA) with defined or putative NADH binding domains. Expression vectors containing these candidate genes were constructed in pTrc99A (pLOI4320, pLOI4317, pLOI4855, and pLOI4856). Amplified regions included the ribosomal binding site, open reading frame, and terminator region. Cell lysates of plasmid-containing strains (LY180) were assayed as previously described (27) for furan reductase (furfural and 5-HMF). None appeared to encode a furan reductase. Activities in all were low and similar to that of the vector control (<0.02 U mg protein−1 for NADH and <0.10 U mg protein−1 for NADPH).
Derivatives of LY180 containing these plasmids were tested for furfural tolerance with (0.1 mM IPTG [isopropyl-β-d-thiogalactopyranoside]) and without IPTG induction using an MIC assay (18, 19, 27). Only pLOI4856 (ucpA) was beneficial (Fig. 1A), increasing the MIC of furfural by 50% (15 mM) compared to that of the vector control and the three other constructs (10 mM). IPTG provided little further benefit, indicating that high levels of UcpA are not needed. Expression of ucpA in LY180(pLOI4856) also increased the MIC for 5-HMF from 16 mM for the control to 20 mM for LY180(pLOI4856) (data not shown).
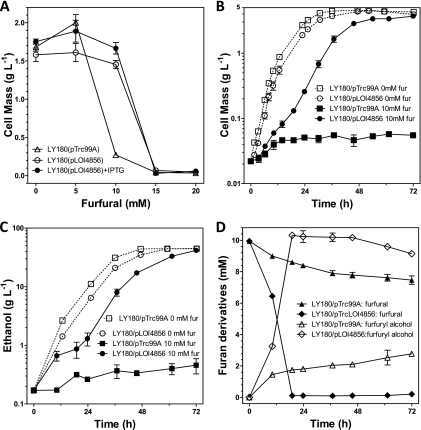
Fig 1.
Plasmid expression of ucpA increases furfural tolerance (MIC) and ethanol production by LY180 (pH-controlled fermentations; 10% xylose). (A) Effect of ucpA plasmid (pLOI4856) on MIC for furfural; (B) effect of pLOI4856 on growth in 10 mM furfural; (C) effect of pLOI4856 on ethanol production in 10 mM furfural; (D) effect of pLOI4856 on furfural metabolism during fermentation. Controls were included without furfural (dotted lines).
The effects of UcpA on growth, ethanol production, and furfural metabolism were investigated in more detail during pH-controlled batch fermentation in mineral salts medium (AM1 medium containing 100 g xylose liter−1, 0.1 mM IPTG and 12.5 μg ml−1 ampicillin for all cultures harboring plasmids, furfural as indicated, and inoculum of 22 mg dry cell weight [dcw] liter−1) as previously described (27). Ethanol (retention time of 1.1 min) and furfuryl alcohol (retention time of 6.2 min) were measured using an Agilent 6890N gas chromatograph (Santa Clara, CA) (18). Furoic acid (retention time of 51.2 min) and sugars were measured by high-performance liquid chromatography (7). Furfural was measured using a Beckman-Coulter DU 800 spectrophotometer (16).
Plasmid pLOI4856 containing ucpA increased furfural tolerance for growth and ethanol production in LY180 compared to those of the control containing pTrc99A (Fig. 1B and C) with IPTG induction. The vector control was substantially inhibited by 10 mM furfural for over 72 h (Table 1), while only a modest initial inhibition was observed for LY180(pLOI4856). During the initial slow phase, LY180(pLOI4856) quantitatively converted furfural to the less toxic furfuryl alcohol (Fig. 1D). No furoic acid was detected. Ethanol production and growth followed similar trends. After furfural was metabolized, the rate of growth and ethanol production increased to near that of controls without furfural, with similar final yields for cell mass and ethanol.
Table 1.
Summary of pH-controlled fermentations (10% xylose)a
| Strain | Furfural concn (mM) | Furfural-dependent slow growth |
μ max (h−1) | Cell yield (g liter−1) | |
|---|---|---|---|---|---|
| μ (h−1) | Duration (h)b | ||||
| LY180/pTrc99A | 0 | No slow phase | 0.23 ± 0.1 | 4.6 ± 0.1 | |
| LY180/pTrc99A | 10 | <0.04 | >72 | <0.04 | 0.06 ± 0.01 |
| LY180/pLOI4856 | 0 | No slow phase | 0.22 ± 0.01 | 4.5 ± 0.2 | |
| LY180/pLOI4856 | 10 | 0.09 ± 0.01 | 22 ± 2 | 0.14 ± 0.01 | 3.6 ± 0.1 |
| LY180 | 0 | No slow phase | 0.23 ± 0.01 | 3.6 ± 0.2 | |
| LY180 | 8 | 0.09 ± 0.01 | 21 ± 1 | 0.16 ± 0.01 | 4.0 ± 0.1 |
| LY180 | 10 | <0.04 | >72 | <0.04 | 0.08 ± 0.02 |
| XW118c | 0 | No slow phase | 0.22 ± 0.03 | 3.1 ± 0.2 | |
| XW118 | 8 | 0.01 ± 0.01 | 55 ± 2 | 0.14 ± 0.01 | 4.0 ± 0.1 |
| XW118 | 10 | <0.04 | >72 | <0.04 | 0.04 ± 0.01 |
| E. coliW | 0 | No slow phase | 0.23 ± 0.01 | 2.0 ± 0.1 | |
| E. coli W | 8 | 0.18 ± 0.01 | 13 ± 1 | 0.19 ± 0.01 | 1.9 ± 0.1 |
| E. coli W | 10 | 0.06 ± 0.02 | 21 ± 1 | 0.20 ± 0.02 | 2.0 ± 0.1 |
| XW137d | 0 | No slow phase | 0.22 ± 0.01 | 2.0 ± 0.1 | |
| XW137 | 8 | 0.11 ± 0.02 | 16 ± 3 | 0.19 ± 0.01 | 1.9 ± 0.1 |
| XW137 | 10 | 0.02 ± 0.01 | 37 ± 2 | 0.12 ± 0.02 | 1.9 ± 0.1 |
| E. coliW/pTrc99A | 0 | No slow phase | 0.23 ± 0.01 | 2.4 ± 0.2 | |
| E. coliW/pTrc99A | 10 | 0.03 ± 0.01 | 26 ± 4 | 0.13 ± 0.01 | 2.0 ± 0.3 |
| E. coli W/pLOI4856 | 0 | No slow phase | 0.22 ± 0.01 | 1.9 ± 0.1 | |
| E. coli W/pLOI4856 | 10 | 0.07 ± 0.01 | 16 ± 2 | 0.18 ± 0.02 | 2.5 ± 0.3 |
Fermentations (n ≥ 2) were performed in AM1 medium with 100 g xylose liter−1 (27).
The duration of furfural-induced slow growth was estimated as the time of intersection using extrapolated rates.
XW118 (LY180 ucpA::kan).
XW137 (E. coli W ucpA::kan).
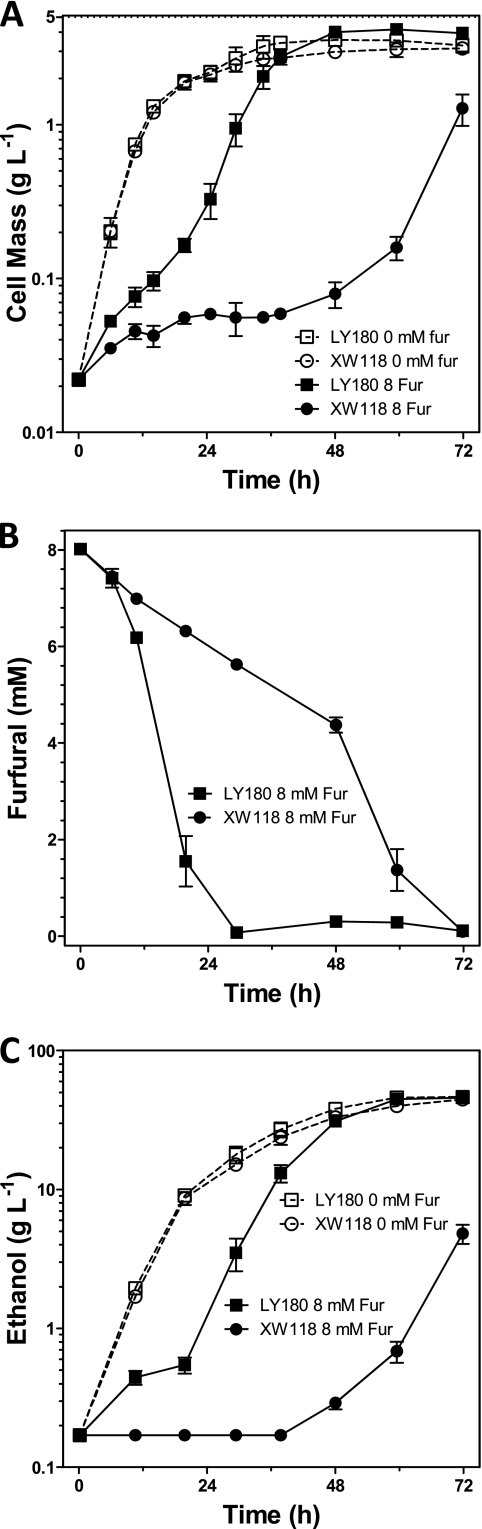
UcpA appears to increase growth in the presence of furfural but does not directly metabolize furfural using NADH or NADPH as electron donors. Although the volumetric rate of furfural reduction was increased by plasmid copies of ucpA (Fig. 1D) whole-cell-specific activity (furfural reductase) (27) was similar to that of the vector control (<0.10 U mg dcw−1). Deletion of chromosomal ucpA in an LY180 background (strain XW118) using Red recombinase technology (5) (Gene Bridges GmbH, Dresden, Germany) decreased furfural tolerance (Fig. 2 and Table 1), confirming that the chromosomally encoded UcpA is functional and beneficial.
Fig 2.
Deletion of chromosomal ucpA (XW118) decreased furfural tolerance of LY180 during pH-controlled fermentations (10% xylose). (A) Cell mass; (B) furfural metabolism; (C) ethanol. Controls were included without furfural (dotted lines).
Strain LY180 has been highly engineered for ethanol production and contains many mutations. Although this engineered strain was more sensitive to inhibition by furfural than the parent strain W, cell yields for LY180 were twice that of strain W with 0 mM and 8 mM furfural (Table 1). Both LY180 and strain W exhibited similar changes in furfural tolerance with regard to ucpA. The addition of plasmid pLOI4856 increased furfural tolerance in strain W (Table 1). Deletion of ucpA from strain W (strain XW137) lowered furfural tolerance. The furfural sensitivity of LY180 may be related to higher aldehyde levels in this homoethanol producer than in strain W (mixed acid fermentation). Mixtures of acetaldehyde and furfural were previously shown to exhibit more than additive toxicity for ethanologenic E. coli (29).
Very little is known about the ucpA gene other than its location, upstream from the cysP operon. UcpA homologues contain a proposed NAD-binding site with homology to short-chain alcohol dehydrogenases (24) and to human 3-hydroxybutyrate dehydrogenase (DHRS6) (8). Reed et al. (23) predicted that this gene may encode diacetyl reductase (acetoin dehydrogenase). These compounds (acetoin, diacetyl, 3-hydroxybutyrate, and acetoacetate) were tested as potential substrates using appropriate cofactors as described by Guo et al. (8). Additional alcohols and aldehydes (ethanol, glycerol, n-butanol, 2-propanol, methanol, 1,3-propanediol, methyglyoxal, dihydroxyacetone, acetaldehyde, butyraldehyde, malondialdehyde, and acrolein) were also tested for alcohol dehydrogenase or aldehyde reductase activities (22, 25). Although an IPTG-induced band corresponding to the predicted size for UcpA (28 kDa) was clearly evident (data not included), lysates of induced LY180(pLOI4856) did not metabolize any of the substrates at a higher rate than control lysates from LY180(pTrc99A).
The mechanism of UcpA action remains unknown. UcpA does not directly metabolize furfural using NADH or NADPH as electron donors. In both LY180 and the parent strain W, furfural retarded fermentation by delaying growth until metabolism to the alcohol form was near completion. UcpA appears to partially restore growth and thereby decrease the time required to complete furfural metabolism. Growth (and fermentation) then resumes at near control rates and final yields for ethanol and cell mass.
Plasmid expression of ucpA was beneficial for both the native W strain and ethanologenic strain LY180. Homologues of UcpA are widely distributed in nature (8, 24) and may be generally useful to improve the furan tolerance in many microbial biocatalysts. Deletion of the chromosomal ucpA was detrimental for furfural tolerance, providing a clear phenotype for this cryptic gene.
Microarray data accession number.
These new microarray data were deposited in the Gene Expression Omnibus (GEO) database at http://ncbi.nlm.nih.gov/geo (accession number GSE34956).
ACKNOWLEDGMENTS
L. O. Ingram is a consultant for Myriant Corp. and a minor shareholder.
This research was supported by the Myriant Corp., the Department of Energy (grant no. DE-FG3608GO88142), and the Department of Agriculture (grant no. 2011-10006-30358).
Footnotes
Published ahead of print 20 January 2012
REFERENCES
- 1.Almeida JR, Bertilsson M, Gorwa-Grauslund MF, Gorsich S, Liden G. 2009. Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 82:625–638 [DOI] [PubMed] [Google Scholar]
- 2.Almeida JR, et al. 2008. NADH- vs NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 78:939–945 [DOI] [PubMed] [Google Scholar]
- 3.Alvira P, Tomas-Pejo E, Ballesteros M, Negro MJ. 2010. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour. Technol. 101:4851–4861 [DOI] [PubMed] [Google Scholar]
- 4.Boronat A, Aguilar J. 1981. Metabolism of l-fucose and l-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J. Bacteriol. 147:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Graef MR, Alexeeva S, Snoep JL, Teixeira de Mattos MJ. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geddes CC, et al. 2011. Simplified process for ethanol production from sugarcane bagasse using hydrolysate-resistant Escherichia coli strain MM160. Bioresour. Technol. 102:2702–2711 [DOI] [PubMed] [Google Scholar]
- 8.Guo K, et al. 2006. Characterization of human DHRS6, an orphan short-chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-beta-hydroxybutyrate dehydrogenase. J. Biol. Chem. 281:10291–10297 [DOI] [PubMed] [Google Scholar]
- 9.Laadan B, Almeida JR, Radstrom P, Hahn-Hagerdal B, Gorwa-Grauslund M. 2008. Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25:191–198 [DOI] [PubMed] [Google Scholar]
- 10.Liu ZL. 2006. Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl. Microbiol. Biotechnol. 73:27–36 [DOI] [PubMed] [Google Scholar]
- 11.Liu ZL, Blaschek HP. 2010. Biomass conversion inhibitors and in situ detoxification, p 233–259. In Vertès A, Oureshi A, Blaschek H, Yukawa H. (ed), Biomass to biofuels: strategies for global industries. John Wiley and Sons, West Sussex, United Kingdom. [Google Scholar]
- 12.Liu ZL, Moon J. 2009. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10 [DOI] [PubMed] [Google Scholar]
- 13.Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S. 2008. Multiple gene mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:743–753 [DOI] [PubMed] [Google Scholar]
- 14.Martinez A, et al. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol. Prog. 17:287–293 [DOI] [PubMed] [Google Scholar]
- 15.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Effects of Ca(OH)2 treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol. Bioeng. 69:526–536 [DOI] [PubMed] [Google Scholar]
- 16.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Use of UV absorbance to monitor furans in dilute acid hydrolysates of biomass. Biotechnol. Prog. 16:637–641 [DOI] [PubMed] [Google Scholar]
- 17.Miller EN, et al. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl. Environ. Microbiol. 75:6132–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller EN, et al. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural resistant ethanologenic Escherichia coli. Appl. Environ. Microbiol. 75:4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller EN, Turner PC, Jarboe LR, Ingram LO. 2010. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol. Lett. 32:661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills TY, Sandoval NR, Gill RT. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22.Pérez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC. 2008. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283:7346–7353 [DOI] [PubMed] [Google Scholar]
- 23.Reed JL, Vo TD, Schilling CH, Palsson BO. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4(9):R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirko A, Wegleńska A, Hryniewicz M, Hulanicka DM. 1997. Characterization of the Escherichia coli gene encoding a new member of the short-chain dehydrogenase/reductase (SDR) family. Acta Biochim. Pol. 44:153–157 [PubMed] [Google Scholar]
- 25.Sulzenbacher G, et al. 2004. Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 342:489–502 [DOI] [PubMed] [Google Scholar]
- 26.Turner PC, et al. 2011. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J. Ind. Microbiol. Biotechnol. 38:431–439 [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. 2011. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl. Environ. Microbiol. 77:5132–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaldivar J, Martinez A, Ingram LO. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524–530 [DOI] [PubMed] [Google Scholar]
- 29.Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65:24–33 [DOI] [PubMed] [Google Scholar]