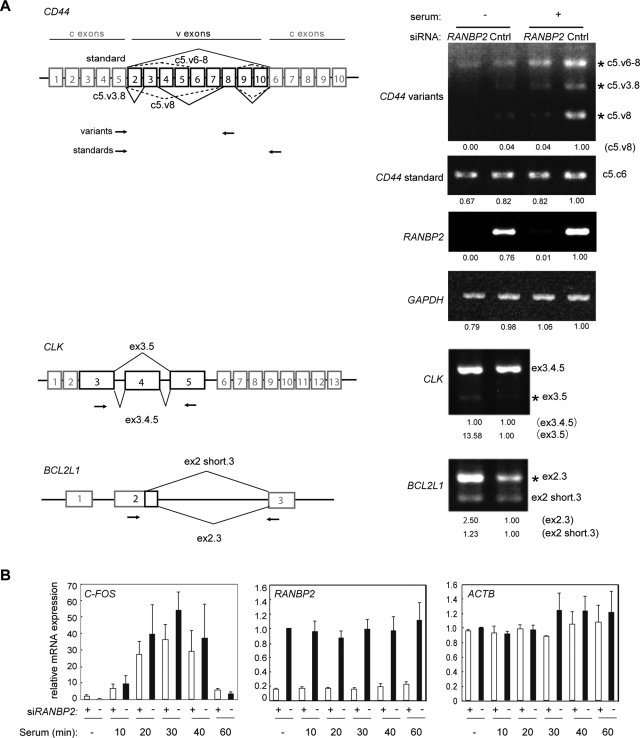
FIGURE 6:
RANBP2 knockdown affects pre-mRNA alternative splicing. (A) Alternative splicing patterns reported for the CD44 (Weg-Remers et al., 2001; Cheng et al., 2006), CLK, and BCL2L1 (Boise et al., 1993) transcripts (left). Variable exons that participate in alternative splicing (v) are indicated by black boxes, and constitutive exons (c) are indicated by gray boxes. Primers used for RT-PCR are shown by arrows. For CD44, the upper and lower pairs of primers amplified the variable and standard isoforms, respectively. HeLa cells were treated with siRNA for 48 h, and total RNA was purified to analyze the splicing patterns (right). For CD44, cells were also serum starved for 24 h prior to stimulation with 20% serum for 4 h. The quantitative band intensity relative to the control is indicated at the bottom. Alternatively spliced products affected by RANBP2 knockdown are marked with asterisks. RANBP2 and GAPDH were used to confirm the knockdown efficiency of RANBP2 and unaffected transcription, respectively. (B) Quantitative RT-PCR of the C-FOS gene under serum induction. The expression of C-FOS, an archetypal immediate early gene, was unaffected by RANBP2 knockdown. RANBP2 and ACTB served as controls for the knockdown efficiency and constant transcription, respectively. The values are the means and SDs of three independent experiments.