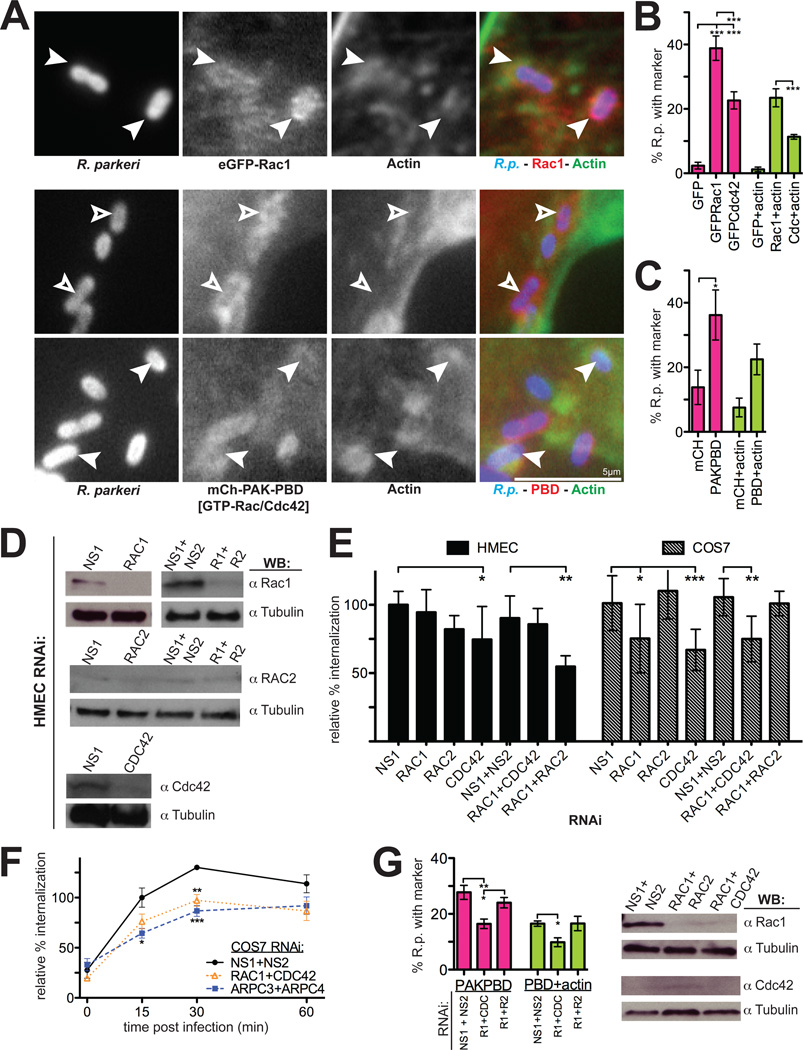
Figure 3. Rho-family GTPases are recruited to invading R. parkeri in mammalian cells and are important for invasion.
(A) R. parkeri (left, blue in merge) are surrounded by eGFP-Rac1 or PAK1-PBD-mCherry (middle, red in merge) and actin (right, green in merge) in HMEC-1 cells at 10 min post-infection. Open arrowheads, Rickettsia associated with PAK1-PBD alone; filled arrowheads, Rickettsia surrounded by actin and eGFP-Rac1 or PAK1-PBD. (B) Percentage of Rickettsia associated with GFP, eGFP-Rac1, eGFP-Cdc42 and actin 10 min post-infection. Pink bars, mean association with marker; light green bars, mean association with both GFP-marker and actin. (C) Percentage of Rickettsia associated with mCherry, PAK1-PBD-mCherry, and actin as in panel (B). (D) Western blots of HMEC-1 cell lysates 48 h post-transfection with the indicated siRNAs, corresponding to one experiment represented in (E). COS-7 cell Western blots are shown in Fig S2A. (E) Relative percent internalization at 15 min post-infection with R. parkeri in either HMEC-1 (solid bars) or COS-7 cells (hashed bars), normalized to nonspecific-RNA treated cells. (F) Relative percentage internalization in COS-7 cells from 0–60 min post-infection with R. parkeri, normalized to 15 min nonspecific-RNA treated cells. (G) Percentage of Rickettsia associated with PAK1-PBD-mCherry and actin after cotransfection with indicated siRNAs, as in (C), with corresponding Western blots. Abbreviations: NS1, nonspecific control siRNA 1; NS2, nonspecific control siRNA 2; R1 + R2, Rac1 and Rac2. Error bars, SEM (B–C, F–G) and SD (E). * p<0.05, ** p<0.01, *** p<0.001 versus indicated control by unpaired Student’s t-test (E–F) or ANOVA with Bonferroni’s post-test (B,C,G). (B–C, F–G) are the mean of two independent experiments performed in duplicate. (E) are the mean of at least three independent experiments performed in duplicate.