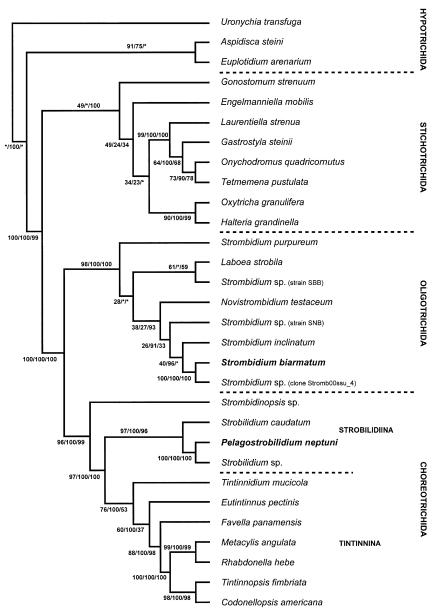
Fig. 8.
The most parsimonious tree inferred from small subunit rRNA gene sequences of spirotrich ciliates and computed with PAUP* ver. 4.0b10 (Swofford 2002). The numbers on the branches are support values for the internal nodes: The first value represents the bootstrap support for the MP analysis (percentage out of 1000 replicates); the second value gives the posterior probability out of 15,001 trees from an ML analysis performed with MrBayes ver. 2.01, employing the Bayesian Inference (Huelsenbeck and Ronquist 2001). The third number is the bootstrap value for the NJ tree (Saitou and Nei 1987), implemented in Phylip ver. 3.6a2 (Felsenstein 1993). Asterisks mark nodes with support values below 12%.