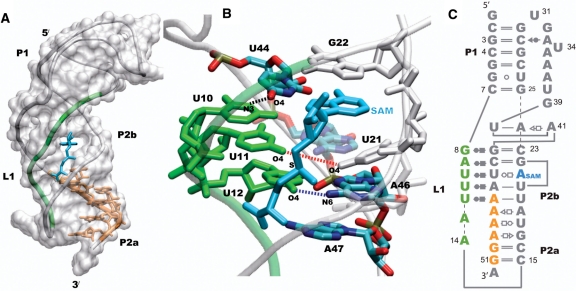
Figure 1.
The classic H-type pseudoknot structure of the SAM-II riboswitch (PDB id 2qwy). L1 (green) is localized in the groove of the P2b helix, and the expression platform which follows the curvature of the P2a/b helix is part of the purine rich sequence shown in orange (A). SAM (cyan) is bound in the groove formed by L1 and the 5′-end of P2b, and the adenine moiety of SAM mimics a base on the 5′-end of P2b by intercalating and stacking between U21 and G22 (B). The secondary structure of the SAM-II riboswitch is shown (C), with the base pairing interactions represented using commonly used notations. The other non-standard interactions observed in the X-ray structure that form the remaining tertiary interactions are not shown. The base-pairing interactions between L1, P2b and SAM in the metabolite bound X-ray crystal structure of the SAM-II riboswitch are shown in (B). The distances between N3 of U10 and O4 of U44 (black), O4 of U11 and O4 of U21 (red), and O4 of U12 and N6 of A46 (blue) are used to monitor the interaction of L1 with the major groove of the helix.