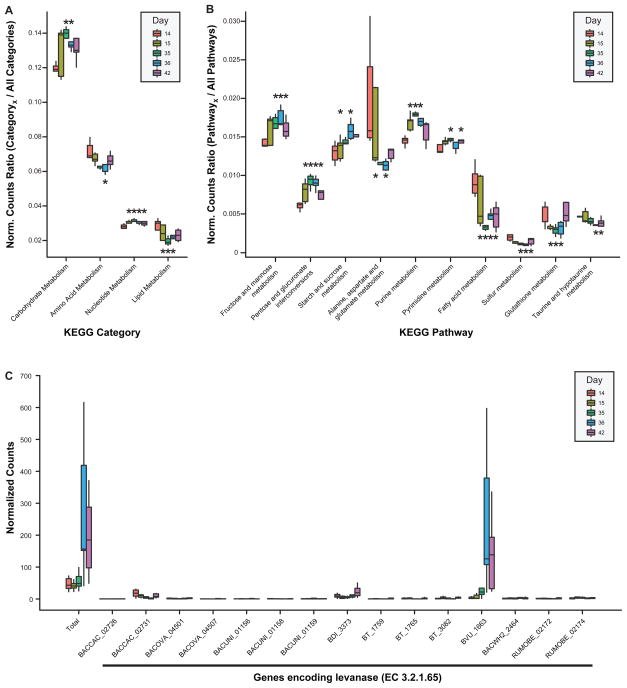
Figure 4. ‘Top-down’ analysis of the effects of the FMP strain consortium on the model 15-member community’s metatranscriptome.
RNA-Seq reads were mapped to the sequenced genomes of the 15 community members. Transcript counts were normalized [reads per kb of gene length per million reads (RPKM), see Supplementary Material] and binned using the hierarchical levels of functional annotation employed by KEGG. For each KEGG category (A) or pathway (B) shown, boxplots depict the proportion of normalized read counts assignable to that annotation out of all reads which could be assigned annotations for that hierarchical level. Data shown correspond to the ‘multiple’ treatment group of mice (the group for which the most time points were collected), however, data for all mice are provided in Table S8. (C) Illustration of how a model community’s functional response (e.g., the increased expression of levanase-encoding genes) can be dissected to identify the subset of genes/species driving the response. Boxes denote top quartile, median, and bottom quartile. Whisker length represents 1.5x inter-quartile range (IQR), except where there are no outliers; in these situations, whiskers span the range from minimum to maximum values. Box color denotes the day fecal samples were obtained (day 14 is the pre-treatment timepoint immediately preceding gavage of the FMP strain consortium). When an asterisk is centered over a box, it indicates that there was a statistically significant change following administration of the FMP consortium relative to the pre-treatment timepoint (p<0.05 by paired, two-tailed Student’s t-test). The positioning of asterisks above versus below a box emphasizes the direction of change (above, upregulation; below, downregulation).