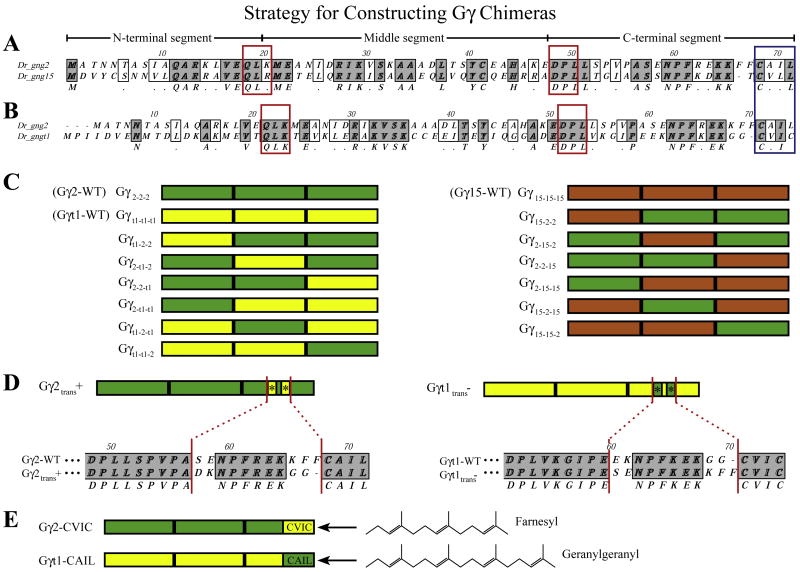
Fig. 1.
Strategy for constructing Gγ subunit chimeras. The gng2 and gng15 (A) or gng2 and gngt1 (B) subunits were aligned with Clustal to show their similarities and differences at the amino acid level. Gγ subunits were split at the conserved sites boxed in red (QLK which corresponds to residues 18–20 in gng2 and DPL, which corresponds to residues 48050 of gng2). The residues that make up the CaaX motif (blue box), and the N-terminal, middle and C-terminal sections have been indicated. The three sections of each gamma subunit were individually PCR amplified and recombined to form the chimeras outlined in (C). Chimeras were named according to the makeup of their three sections as indicated to the left of each subunit Swapping the ‘translocation to endomembranes’ motif (*) involved mutating the five amino acids indicated in an alignment of their C-terminal sequences (D). Subunits harboring these point mutations were named according to whether their new motif should (trans+) or should not (trans-) be consistent with the ability to translocate upon receptor activation. Subunits with swapped CaaX motifs have been named with the amino acids that make up their new motif (E) (gng2-WT terminates with -CAIL; after swap = gng2-CVIC). The prenyl lipid that will be post-translationally added to these Gγ chimeras is indicated to the right.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)