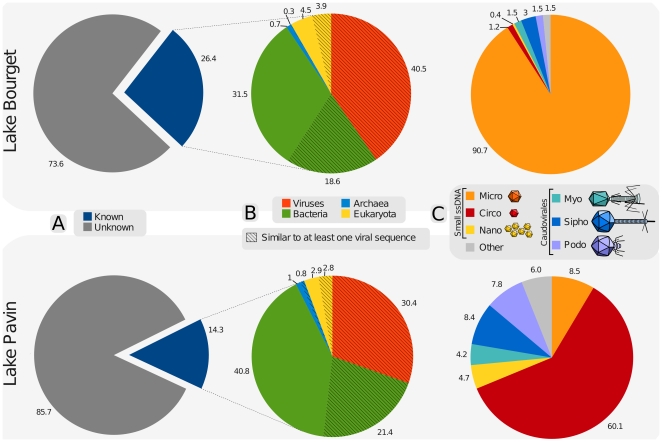
Figure 1. Composition and taxonomic affiliations of Lake Bourget and Lake Pavin virome reads as determined by similarity to known sequences.
(A) The percent of “known” virome sequences when compared to the NR protein database. A read was considered “known” if it had a significant similarity in NR (BLASTx using thresholds of 10−3 on e-value and 50 on bit score). (B) Breakdown of the “known” sequences into Viruses, Bacteria, Archaea, or Eukarya using similarity results against NR. Hatched parts were reads having a best BLAST hit against a non-viral sequence, but still presenting significant similarity against a complete virus genome sequence of the RefseqVirus database (tBLASTx using thresholds of 10−3 on e-value and 50 on bit score) and thus designated as reads “similar to at least one viral sequence”. (C) Taxonomic composition at the viral family level of these reads “similar to at least one viral sequence” computed using the GAAS pipeline. The “Other” category pools families which represented less than 1% of the full virome sequences. The number of sequences represented in each chart are as follow : 593,084 ; 156 772 ; 95,905 for charts A, B and C of Lake Bourget virome, and 649,290 ; 92,834 ; 47,345 for the Lake Pavin virome.