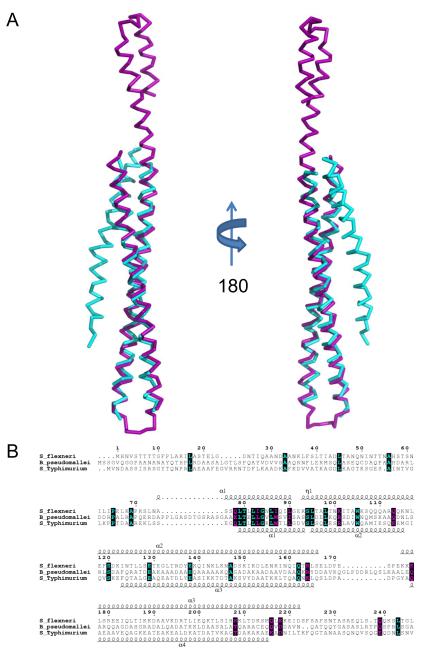
Fig. 3. Structural Superposition of Translocator Coiled-coils.
A, Ribbon diagram of a structural alignment of the coiled-coils from IpaB (residues 120-224, purple) and SipB (residues 126-226, cyan) with an RMSD of 1.42 Å over 93/94 Cα atoms within 5.0 Å, rotated 180° about the long axis. Although the overall topology of both structures is similar, there are differences within the N-terminal region spanning the first helix (α1) and turn as well as the length of the second helix (α2). Such differences within the N-terminus of the structures reported here could be reflective of the apparent instability of the chaperone binding domains (CBD) in the absence of their cognate chaperones. B, Limited structure-based sequence alignment of type III secretion first translocators (residues 1-240) colored according to residue conservation (cyan=absolute and purple=similar) as judged by the BLOSUM62 matrix. Alignment was generated using ClustalW and rendered with ESPRIPT. Numbers above the sequences correspond to S. flexneri IpaB. Secondary structure elements of IpaB and SipB are shown above and below the alignment, respectively. Representations of all structures were generated using PyMol 46. Three-dimensional structures were superimposed using the Local-Global Alignment method (LGA) 47. Sequence alignments were carried out using CLUSTALW 48 and aligned with secondary structure elements using ESPRIPT 49.