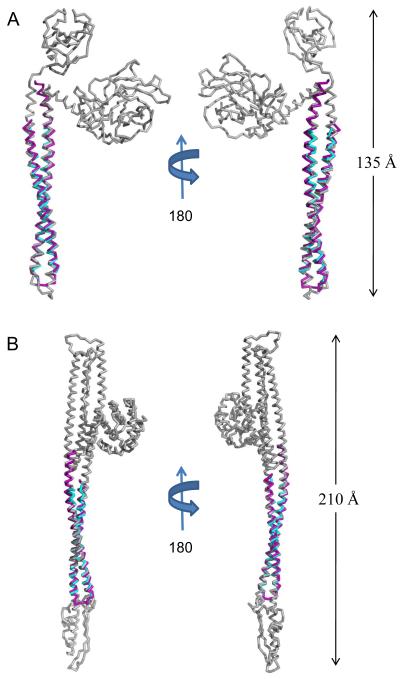
Fig. 4. Translocator Coiled-coils Share Structural Homology with Pore-forming Proteins.
A, Ribbon diagram of IpaB (residues 104-224, purple) and SipB (residues 126-226, cyan) superimposed upon E. coli Colicin E3 (colored grey; PDB code= 1JCH) with an RMSD of 2.03 Å over 118/121 Cα atoms for IpaB and an RMSD of 1.19 Å over 92/94 Cα atoms for SipB. Crystal structures are rotated 180° about their long axis. B, Ribbon diagram of IpaB (residues 104-224, purple) and SipB (residues 126-226, cyan) superimposed over E. coli Colicin Ia (colored grey; PDB code= 1CII) with an RMSD of 1.73 Å over 121/121 Cα atoms for IpaB and an RMSD of 1.21 Å over 93/94 Cα atoms for SipB. Crystal structures are rotated 180° about their long axis. The DALI server was used to query available structures within the PDB for structural homology 35. Representations of all structures were generated using PyMol 46. Three-dimensional structures were superimposed using the Local-Global Alignment method (LGA) 47.