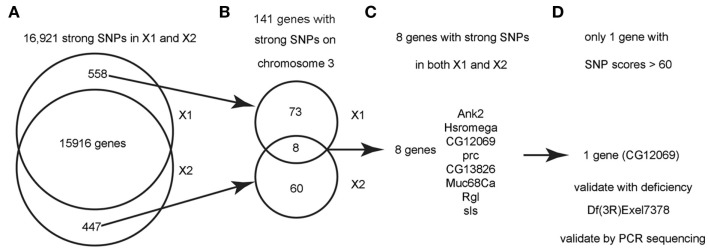
Figure 3.
Flowchart for finding the causative SNPs in X1 and X2. (A) SnpEeff identified 16,921 “class 1” SNPs (see text) with a quality score > 1 in both X1 and X2 (zero quality scores are usually resulted from reads mapping to multiple genomic regions). There are 558 SNPs that are only present in X1 and 447 SNPs that are only present in X2. (B) Since we know that X1 and X2 are on chromosome 3, we focused on the 141 strong SNPs on chromosome 3 that are present in X1 or X2 but not both. There are only eight genes that are commonly affected by unique SNPs in both X1 and X2 (note that the eight genes have at least two SNPs at different bases). (C) List of the eight genes with SNPs in both X1 and X2. See Table 1 for more details. (D) Only one gene, CG12069/Pka-like, contained SNPs with scores > 60. These SNPs were validated by capillary sequencing of PCR-amplified DNA from the genetic interval of the male-sterile locus as defined by meiotic and deletion mapping data (see text). ca.