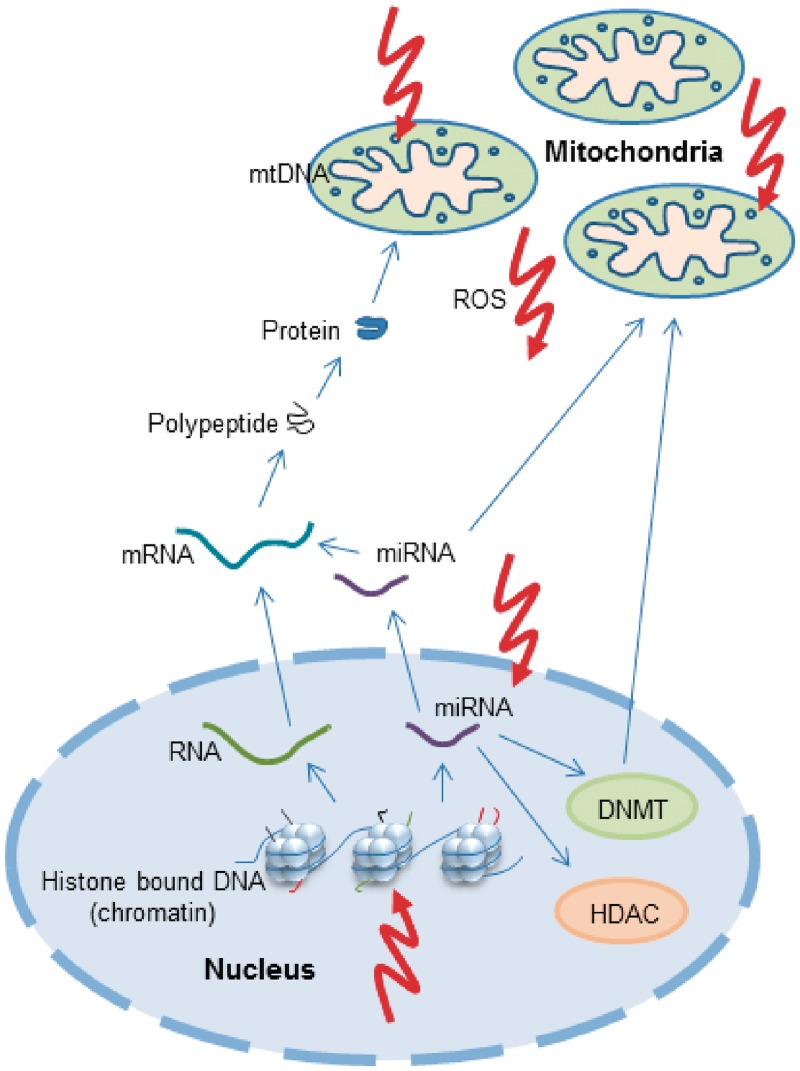
Figure 1.
A schematic representation of mitochondrial and nuclear genomes and their inter-relation with epigenetic factors. The nuclear genome is coiled around histone octamers to form nucleosomes. The tails of histone proteins are decorated with a variety of modifications that influence the regulation of gene expression. Permissive histone markings allow transcription from DNA to RNA, post-transcription processing to mRNA and translation to polypeptides and thus proteins. Nuclear-encoded mitochondrial proteins are then translocated into the mitochondrion. mtDNA also encodes genes essential for intra-mitochondrial protein synthesis, but this genome is not histone bound. In addition to mRNA, miRNA are also transcribed from nuclear DNA and can interfere with mRNA to induce degradation or suppress translation. miRNAs can influence mitochondrial metabolism and some miRNAs are known to directly activate the generation of ROS. Furthermore, miRNAs influence the expression of DNMT and HDAC enzymes. ROS produced by mitochondria or other endogenous sources can damage the mtDNA genome directly as well as influence epigenetic machinery at several levels, either through damage to miRNA or through the alteration of histone modifications. DNMTs translocate to the mitochondria and bind to mtDNA, although evidence that this is to effect epigenetic regulation remains elusive. HDAC, histone deacetylase; mRNA, messenger RNA; ROS, reactive oxygen species