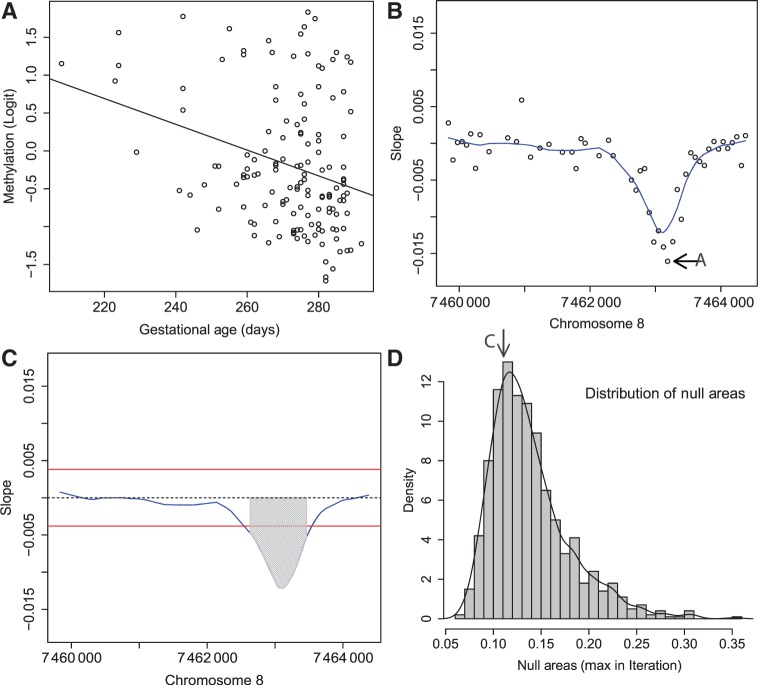
Figure 2.
Step-by-step illustration of our bump-hunting algorithm. (A) Logit-transformed methylation measurements are plotted against the outcome of interest (gestational age) for a specific probe j. A regression line obtained from fitting the model presented in Equation 1 is shown as well. The estimated slope 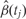 is retained for the next step. (B) For 48 consecutive probes, the estimated
is retained for the next step. (B) For 48 consecutive probes, the estimated 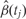 are plotted against their genomic location tj. The specific estimated slope from the probe in (A) is indicated by ‘A’ and an arrow. The blue curve represents the smooth estimate
are plotted against their genomic location tj. The specific estimated slope from the probe in (A) is indicated by ‘A’ and an arrow. The blue curve represents the smooth estimate 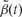 obtained using loess. (C) The smooth estimate
obtained using loess. (C) The smooth estimate 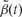 from (b) is shown but here with predefined thresholds represented by red horizontal lines. The region for which
from (b) is shown but here with predefined thresholds represented by red horizontal lines. The region for which 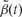 exceeds the lower threshold is considered a candidate DMR. The area shaded in grey is used as a summary statistic. (D) A null distribution for the area summary statistic described in (c) is estimated by performing using permutations (as described in the text). The histogram summarizes the null areas obtained from permutations and estimates the null distribution. The area obtained from the region shown in (C) is highlighted with an arrow and the label ‘C’. Note that this DMR region is not statistically significant as it can easily happen by chance
exceeds the lower threshold is considered a candidate DMR. The area shaded in grey is used as a summary statistic. (D) A null distribution for the area summary statistic described in (c) is estimated by performing using permutations (as described in the text). The histogram summarizes the null areas obtained from permutations and estimates the null distribution. The area obtained from the region shown in (C) is highlighted with an arrow and the label ‘C’. Note that this DMR region is not statistically significant as it can easily happen by chance