Figure 4.
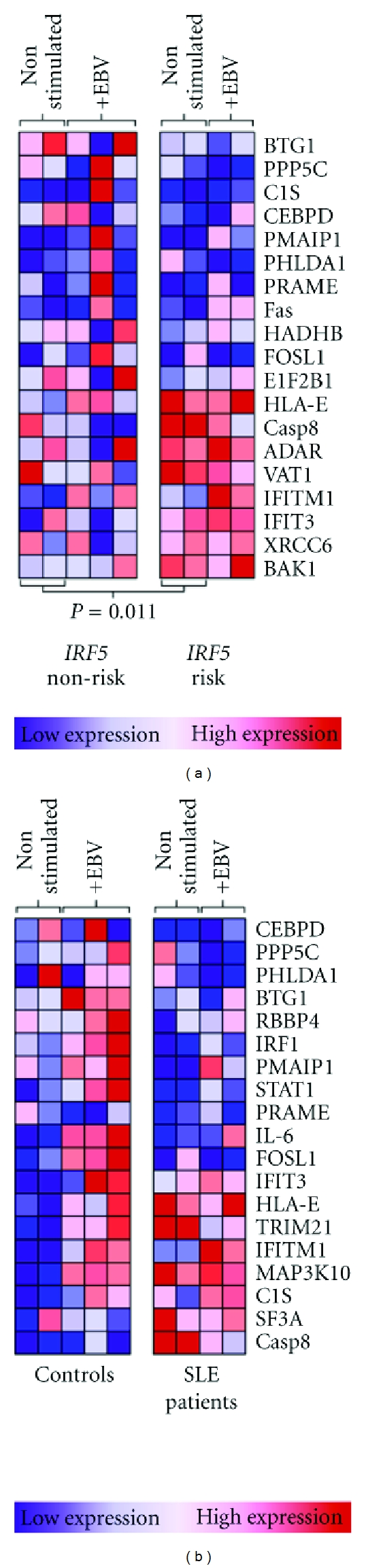
Dependence of the interferon response signature on IRF5 haplotype in SLE patients. Interferon signature genes identified in gene set enrichment analysis were compared between patients and controls, and between lupus patients with the IRF5 risk and nonrisk haplotypes. Each column represents expression of genes of interest in individual subjects. (a) Interferon signature genes of 3 IRF5 nonrisk and 2 IRF5 risk SLE patients. Gene expression comparisons are between IRF5 risk and nonrisk SLE patients, either nonstimulated or EBV infected. (b) Gene set enrichment of 3 control IRF5 risk haplotype individuals and 3 SLE patient IRF5 risk haplotype individuals. Gene expression comparisons between IRF5 risk patients and controls from nonstimulated and EBV-infected B cells. ADAR: adenosine deaminase, BAK: Bcl2-antagonist/killer, BTG: B cell translocation gene, C1S: complement component 1S, CASP: caspase, CEBPD: CCAAT/enhancer-binding protein δ, eIF2B: eukaryotic translation initiation factor 2B, FOSL: Fos-related antigen, HADHB: hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase β subunit, HLA: human leukocyte antigen, PHLDA: pleckstrin homology-like domain family A, PMAIP1: phorbol-12-myristate-13-acetate-induced protein 1 (Noxa), PRAME: melanoma antigen preferentially expressed in tumors, RBBP: retinoblastoma binding protein, SF3A: splicing factor 3A, TRIM: tripartite motif, VAT: vesicle amine transport, and XRCC: X-ray repair cross-complementing (Ku70).
